# 安装包
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("networkD3", quietly = TRUE)) {
install.packages("networkD3")
}
if (!requireNamespace("dplyr", quietly = TRUE)) {
install.packages("dplyr")
}
if (!requireNamespace("readxl", quietly = TRUE)) {
install.packages("readxl")
}
if (!requireNamespace("webshot", quietly = TRUE)) {
install.packages("webshot")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("openxlsx", quietly = TRUE)) {
install.packages("openxlsx")
}
if (!requireNamespace("ggalluvial", quietly = TRUE)) {
install.packages("ggalluvial")
}
# 加载包
library(ggplot2)
library(networkD3)
library(dplyr)
library(readxl)
library(webshot)
library(tidyverse)
library(openxlsx)
library(ggalluvial)桑基图
桑基图允许研究流量。实体(节点)由矩形或文本表示,箭头或弧线用于显示它们之间的流量,宽度与流量的重要性成正比。在 R 中,networkD3 包是构建它们的最佳方式。
示例
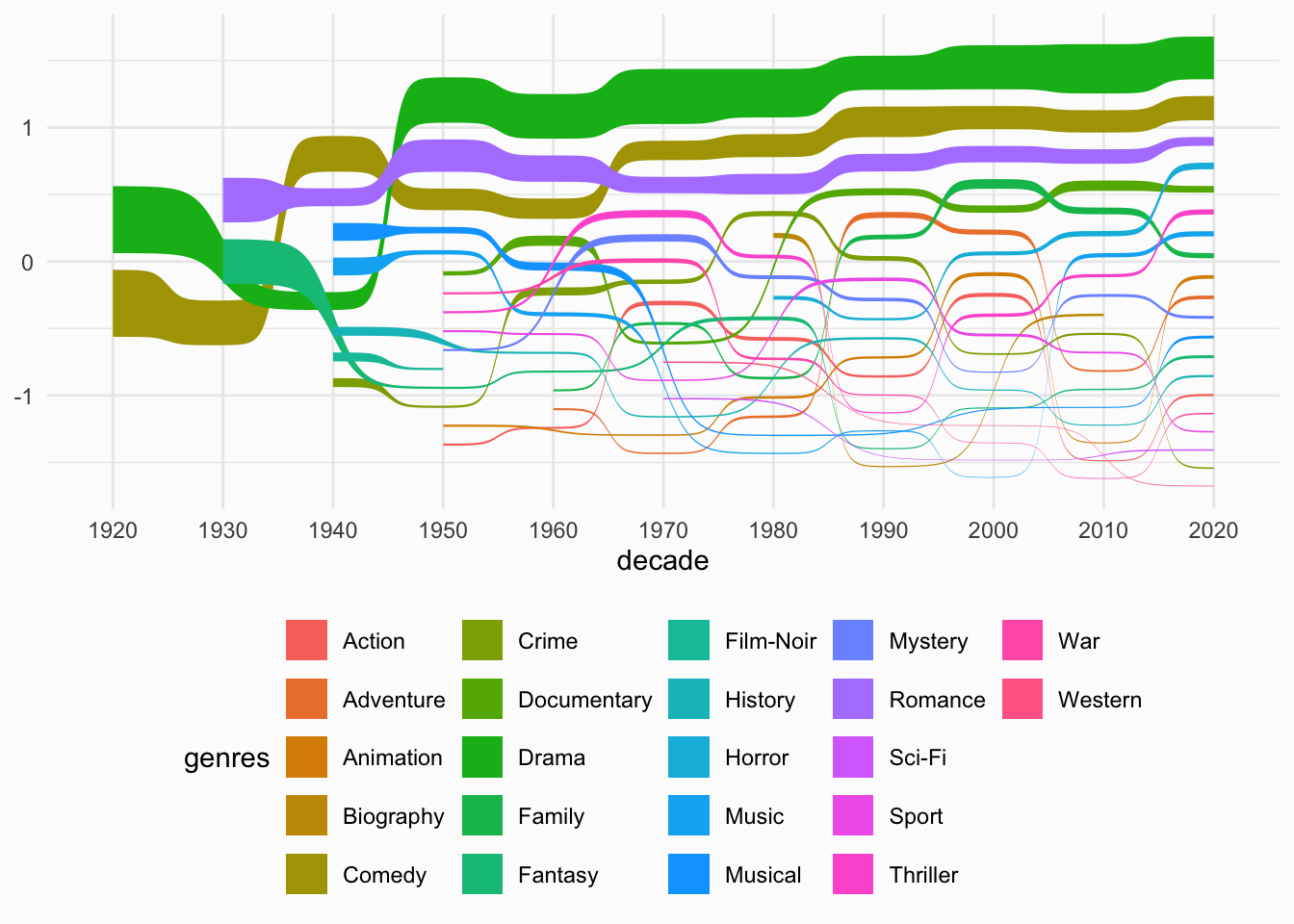
这张图表使用桑基曲线图可视化了过去几十年夏季档电影的类型分布情况。它重点展示了三种最常见的类型(剧情片、喜剧片和爱情片),并显示了它们的比例随时间的变化。
环境配置
系统要求: 跨平台(Linux/MacOS/Windows)
编程语言:R
依赖包:
ggplot2,networkD3,dplyr,readxl,webshot,tidyverse,openxlsx,ggalluvial
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-02
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
dplyr * 1.1.4 2023-11-17 [1] RSPM
forcats * 1.0.1 2025-09-25 [1] RSPM
ggalluvial * 0.12.5 2023-02-22 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
lubridate * 1.9.4 2024-12-08 [1] RSPM
networkD3 * 0.4.1 2025-04-14 [1] RSPM
openxlsx * 4.2.8.1 2025-10-31 [1] RSPM
purrr * 1.2.1 2026-01-09 [1] RSPM
readr * 2.1.6 2025-11-14 [1] RSPM
readxl * 1.4.5 2025-03-07 [1] RSPM
stringr * 1.6.0 2025-11-04 [1] RSPM
tibble * 3.3.1 2026-01-11 [1] RSPM
tidyr * 1.3.2 2025-12-19 [1] RSPM
tidyverse * 2.0.0 2023-02-22 [1] RSPM
webshot * 0.5.5 2023-06-26 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
以下是一个使用临床数据库中某种药物临床数据的简要教程。该数据集研究了药物对患者血糖水平的影响,血糖水平分为三个等级:低血糖(<3.9 mmol/L)、正常血糖(3.9-6.1 mmol/L)和高血糖(>6.1 mmol/L)。药物给药前后血糖水平变化的绝对值用“value”表示。本示例演示了如何加载和处理这些数据集。
# 读取药物临床数据集
drugs <- read.csv("https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/drugs.csv", stringsAsFactors = FALSE)
# 创建一个节点数据框
nodes <- data.frame(
name=c(as.character(drugs$source),
as.character(drugs$target)) %>% unique())
# 重新格式化
drugs$IDsource <- match(drugs$source, nodes$name)-1
drugs$IDtarget <- match(drugs$target, nodes$name)-1
# 读取药物临床数据集
drug <- read.csv("https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/drug.csv", stringsAsFactors = FALSE)
levels(drug$`glucose(mmol/L)`) <- rev(levels(drug$`glucose(mmol/L)`))可视化
1. 可视化
1.1 基础桑基图
图 1 桑基图描述了患者在使用某种药物前后血糖水平的变化。
# 基础桑基图
p1 <- sankeyNetwork(Links = drugs, Nodes = nodes,
Source = "IDsource", Target = "IDtarget",
Value = "value", NodeID = "name")
p11.2 自定义颜色
使用 JavaScript 调用
图 2 桑基图描述了患者在使用某种药物前后血糖水平的变化。
第一步是创建一个用于颜色映射的 JavaScript 对象。然后,为每个节点分配一个颜色。最后,在 networkD3 的 colourScale 参数中调用此对象。
# 准备颜色标度:为每个节点分配一种特定的颜色。
my_color <- 'd3.scaleOrdinal() .domain(["before-normal", "before-high","before-low", "after-high", "after-low", "after-normal"]) .range(["steelblue", "red" , "#69b3a2", "red", "#69b3a2", "steelblue"])'
p2_1 <- sankeyNetwork(Links = drugs, Nodes = nodes,
Source = "IDsource", Target = "IDtarget",
Value = "value", NodeID = "name",
colourScale=my_color,fontSize=15,nodePadding=20,nodeWidth=25)
p2_1图 3 这张桑基图显示了患者在使用某种药物前后血糖水平的变化。蓝色组表示血糖水平变化大于 1 mmol/L,绿色组表示变化小于 1 mmol/L。
# 在每个链接中添加一个“组”列
drugs$group <- as.factor( c("type_a","type_a","type_b","type_b","type_b","type_b","type_a","type_b","type_b","type_b","type_a","type_a","type_a","type_b","type_a","type_a","type_a","type_a"))
# 为每个节点添加一个“分组”列。这里,所有节点都被放在同一个组中,使它们显示为灰色。
nodes$group <- as.factor(c("my_unique_group"))
# 给每个组分配颜色
my_color <- 'd3.scaleOrdinal() .domain(["type_a", "type_b", "my_unique_group"]) .range(["#69b3a2", "steelblue", "grey"])'
p2_3 <- sankeyNetwork(Links = drugs, Nodes = nodes,
Source = "IDsource", Target = "IDtarget",
Value = "value", NodeID = "name",
colourScale=my_color,LinkGroup="group", NodeGroup="group")
p2_32. 使用ggalluvial包
ggalluvial是ggplot2的扩展包,它遵循ggplot2的图层化语法,用于创建冲积图,与桑基图有相似之处,但是它是根据数据和一组参数唯一确定的。
2.1 基础桑基图
图 4 这张桑基图显示了患者在使用某种药物前后血糖水平的变化。
# 基础桑基图
p4_1 <- ggplot(drug,
aes(x = time, stratum = level, alluvium = id,
y = value,
fill = level, label = level)) +
scale_x_discrete(expand = c(.1, .1)) +
geom_flow() +
geom_stratum(alpha = .5) +
geom_text(stat = "stratum", size = 3) +
theme(legend.position = "none")
p4_1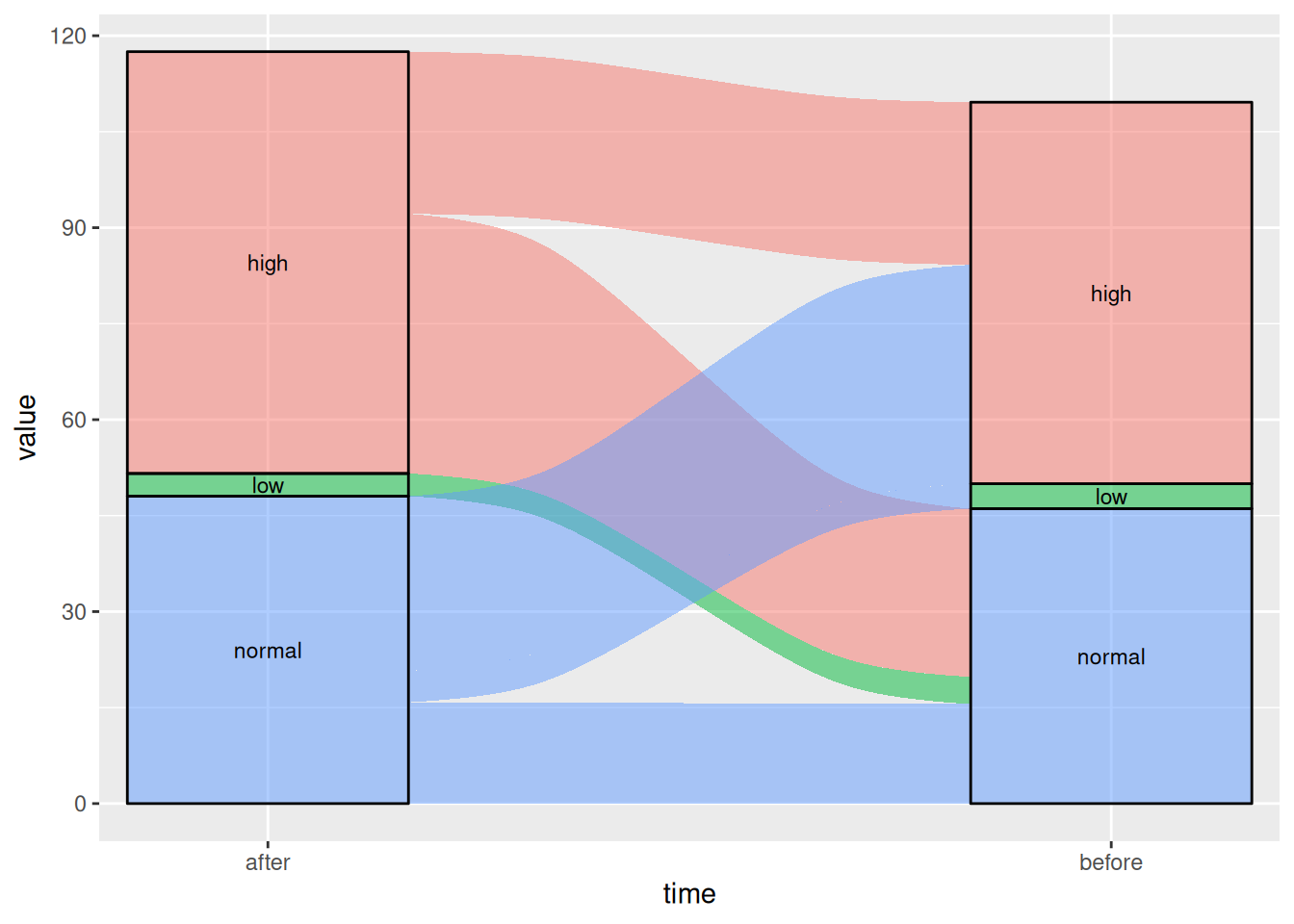
2.2 改变线条类型
通过改变curve_type参数,有”linear”,“cubic”, “quintic”, “sine”,“arctangent”, “sigmoid”. “xspline”7种选项。
图 5 该桑基图显示了患者在使用某种药物前后血糖水平的变化。
# Linear
p5_1 <- ggplot(drug,
aes(x = time, stratum = level, alluvium = id,
y = value,
fill = level, label = level)) +
scale_x_discrete(expand = c(.1, .1)) +
geom_alluvium(curve_type = "linear")+
geom_stratum(alpha = 1) +
geom_text(stat = "stratum", aes(label = after_stat(stratum))) +
theme(legend.position = "none")
p5_1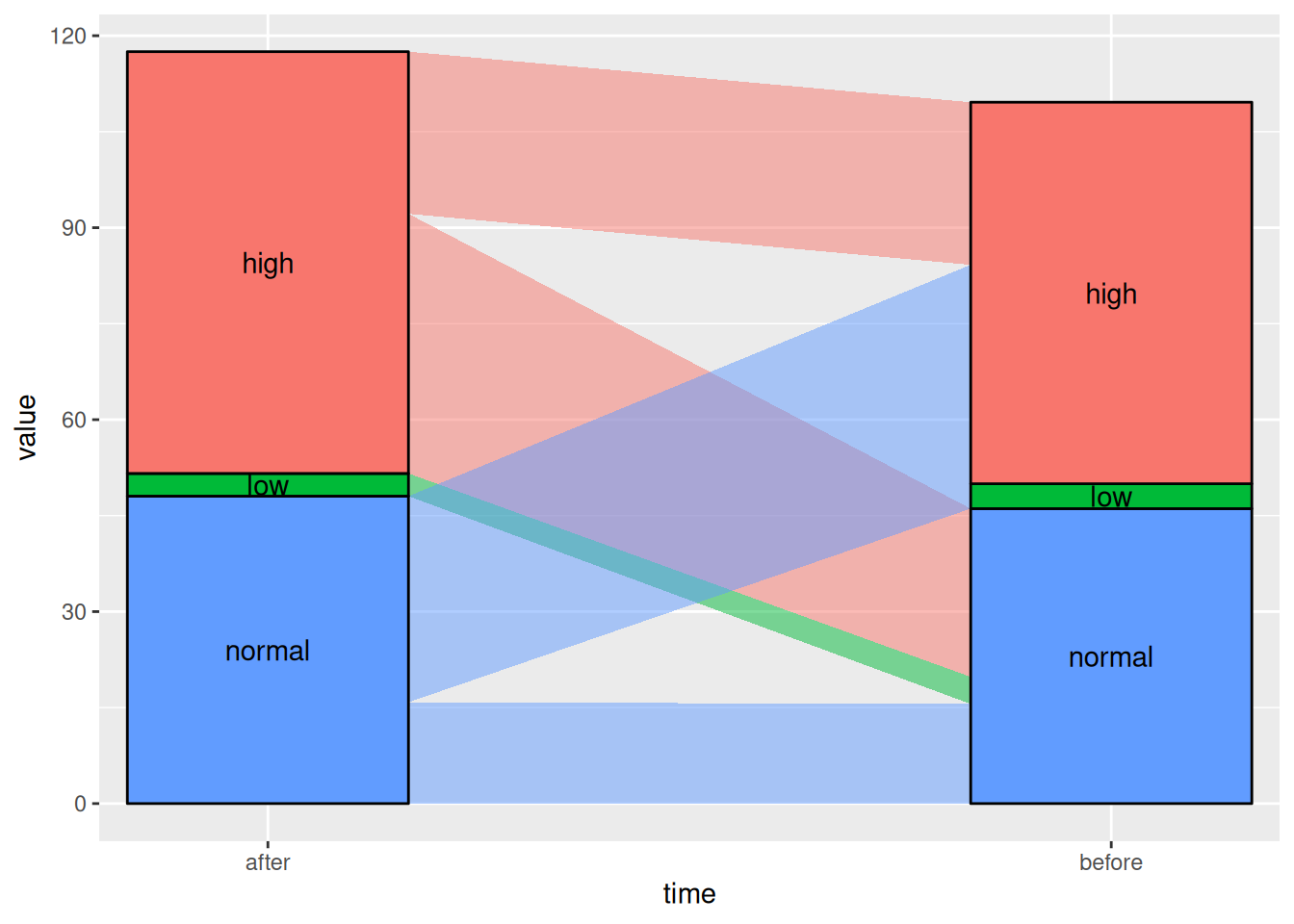
图 6 该桑基图显示了患者在使用某种药物前后血糖水平的变化。
# Sigmoid
p5_2 <- ggplot(drug,
aes(x = time, stratum = level, alluvium = id,
y = value,
fill = level, label = level)) +
scale_x_discrete(expand = c(.1, .1)) +
geom_alluvium(curve_type = "sigmoid")+
geom_stratum(alpha = 1) +
geom_text(stat = "stratum", aes(label = after_stat(stratum))) +
theme(legend.position = "none")
p5_2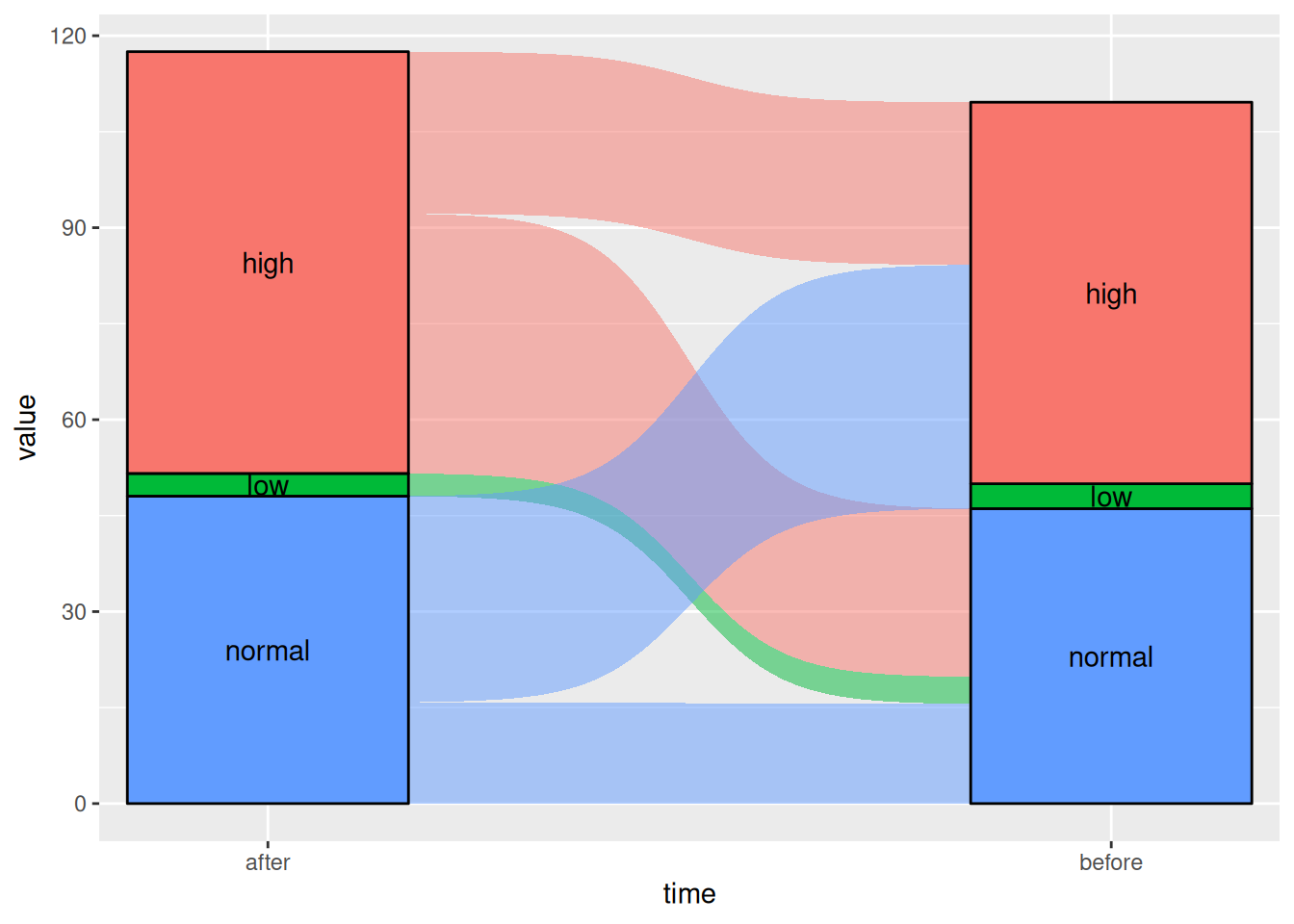
应用场景
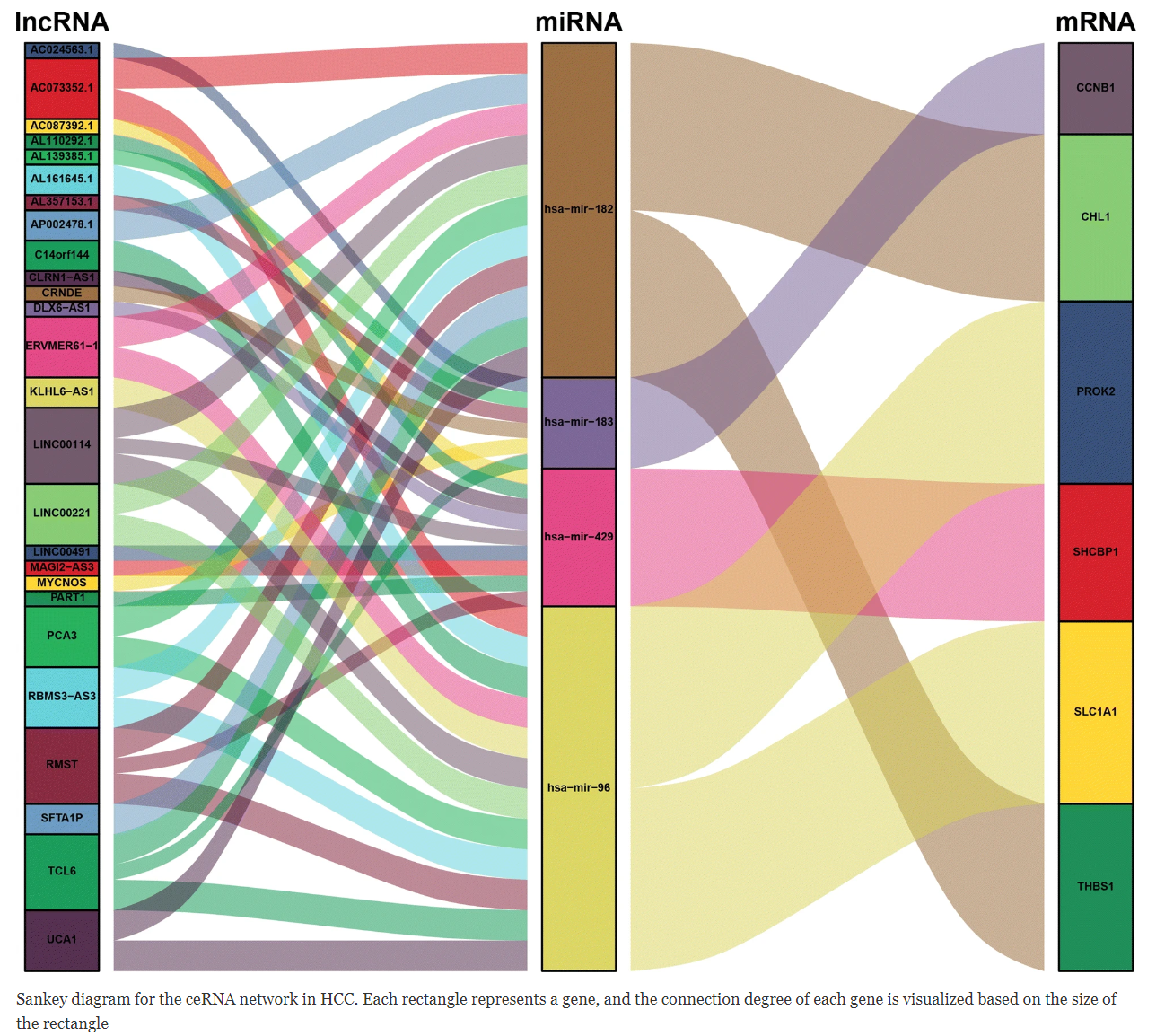
在ceRNA相关的研究中,例如circRNA-miRNA-mRNA,或者lncRNA-miRNA-mRNA的靶向关系图谱,一般通过网络图呈现。 [1]
参考文献
[1] Long J, Bai Y, Yang X, Lin J, Yang X, Wang D, He L, Zheng Y, Zhao H. Construction and comprehensive analysis of a ceRNA network to reveal potential prognostic biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2019 Apr 11;19:90. doi: 10.1186/s12935-019-0817-y. PMID: 31007608; PMCID: PMC6458652.
[2] The R Graph Gallery – Help and inspiration for R charts (r-graph-gallery.com)
[3] Gandrud, Christopher, et al. networkD3: D3 JavaScript Network Graphs from R. Version 0.4, 2017. https://CRAN.R-project.org/package=networkD3.
[4] Wickham, H., & François, R. (2016). dplyr: A Grammar of Data Manipulation [Computer software]. Retrieved from https://CRAN.R-project.org/package=dplyr
[5] Wickham, H. (2009). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. https://ggplot2.tidyverse.org
[6] Allaire, J. J., & Xie, Y. (2018). webshot: Save Web Content as an Image File [Computer software]. Retrieved from https://CRAN.R-project.org/package=webshot
[7] Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L. D., François, R., … Yutani, H. (2019). tidyverse: Easily Install and Load the ‘Tidyverse’ (Version 1.2.1) [Computer software]. Retrieved from https://CRAN.R-project.org/package=tidyverse
[8] Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L. D., François, R., … Yutani, H. (2019). tidyverse: Easily Install and Load the ‘Tidyverse’ (Version 1.2.1) [Computer software]. Retrieved from https://CRAN.R-project.org/package=tidyverse
[9] Kassambara, W. (2020). ggsankey: Create Sankey Diagrams with ‘ggplot2’ [Computer software]. Retrieved from https://CRAN.R-project.org/package=ggsankey
[10] Brunson JC, Read QD. ggalluvial: Alluvial Plots in ‘ggplot2’. R package version 0.12.5. 2023. https://CRAN.R-project.org/package=ggalluvial
