# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("survival", quietly = TRUE)) {
install.packages("survival")
}
if (!requireNamespace("rms", quietly = TRUE)) {
install.packages("rms")
}
if (!requireNamespace("ggplotify", quietly = TRUE)) {
install.packages("ggplotify")
}
# 加载包
library(data.table)
library(jsonlite)
library(survival)
library(rms)
library(ggplotify)诺莫图
注记
Hiplot 网站
本页面为 Hiplot Nomogram 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
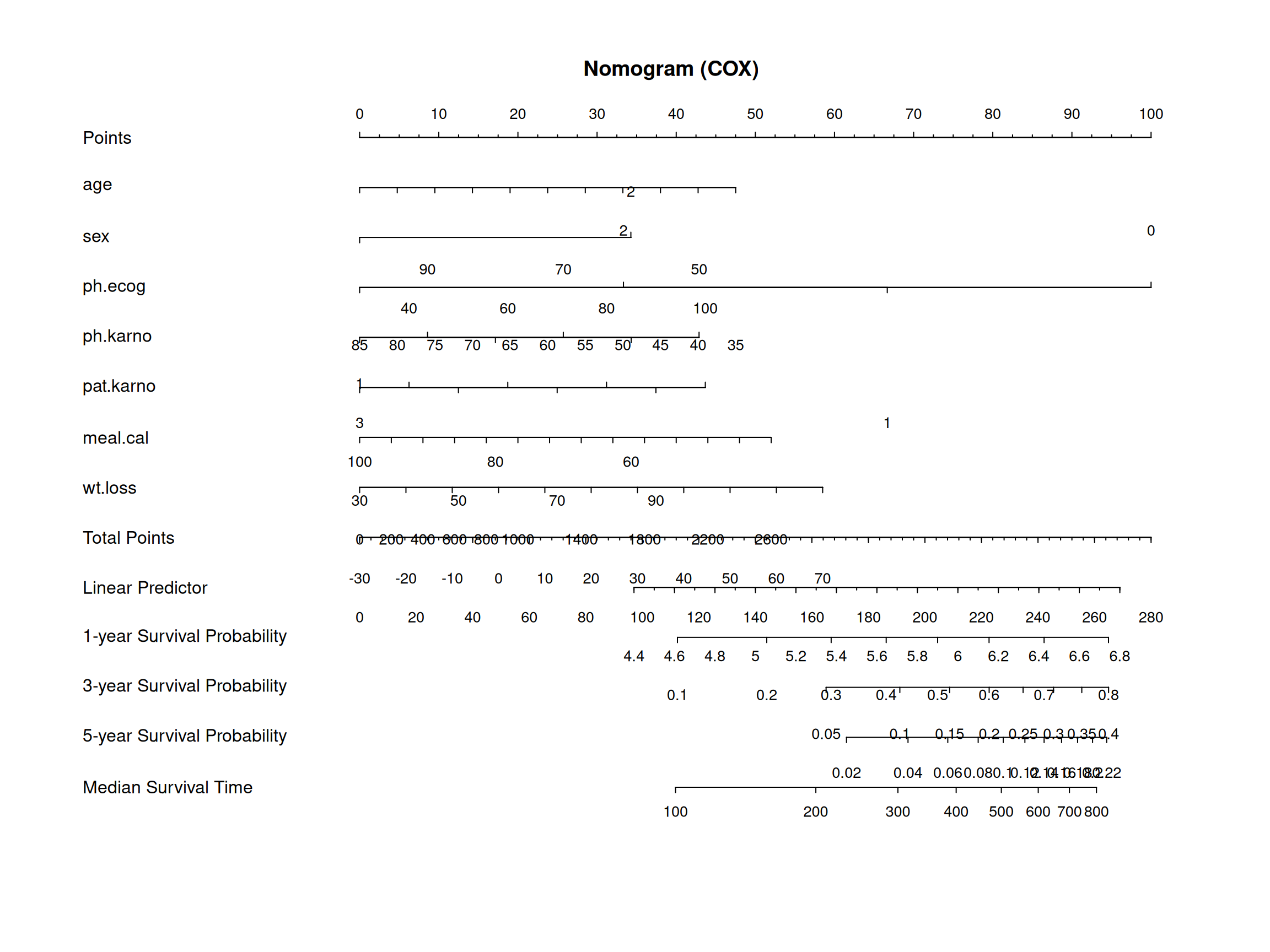
诺莫图常被用来评价肿瘤和医学的预后,并能直观地反映logistic回归或Cox回归的结果。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;survival;rms;ggplotify
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-28
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.0 2025-12-24 [1] RSPM
ggplotify * 0.1.3 2025-09-20 [1] RSPM
Hmisc * 5.2-5 2026-01-09 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
rms * 8.1-0 2025-10-14 [1] RSPM
survival * 3.8-3 2024-12-17 [3] CRAN (R 4.5.2)
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
随时间变化的生存数据帧,根据实例数据用0,1等数字表示性别和状态。
# 加载数据
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/nomogram/data.json")$exampleData$textarea[[1]])
data <- as.data.frame(data)
# 整理数据格式
dd <- datadist(data)
options(datadist = "dd")
## 建立 COX 模型并运行列线图
cox_res <- psm(
data = data,
as.formula(paste(
sprintf("Surv(%s, %s) ~ ", colnames(data)[1], colnames(data)[2]),
paste(colnames(data)[3:length(colnames(data))],
collapse = "+"
)
)),
# Surv(time, status) ~ age + sex + ph.ecog + ph.karno + pat.karno,
dist = "lognormal"
)
## 建立 Survival 概率函数
surv <- Survival(cox_res)
## 构建分位数生存时间函数
med <- Quantile(cox_res)
cox_nomo <- nomogram(
cox_res,
fun = list(function(x) surv(365, x), function(x) surv(1095, x),
function(x) surv(1825, x), function(x) med(lp = x)),
funlabel = c("1-year Survival Probability",
"3-year Survival Probability",
"5-year Survival Probability",
"Median Survival Time"),
maxscale = 100
)
# 查看数据
head(data) time status age sex ph.ecog ph.karno pat.karno meal.cal wt.loss
1 306 2 74 1 1 90 100 1175 NA
2 455 2 68 1 0 90 90 1225 15
3 1010 1 56 1 0 90 90 NA 15
4 210 2 57 1 1 90 60 1150 11
5 883 2 60 1 0 100 90 NA 0
6 1022 1 74 1 1 50 80 513 0可视化
# 诺莫图
p <- as.ggplot(function() {
plot(cox_nomo, scale = 1)
title(main = "Nomogram (COX)")
})
p