# install packages
if (!requireNamespace("survival", quietly = TRUE)) {
install.packages("survival")
}
if (!requireNamespace("survminer", quietly = TRUE)) {
install.packages("survminer")
}
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("dplyr", quietly = TRUE)) {
install.packages("dplyr")
}
if (!requireNamespace("tidyr", quietly = TRUE)) {
install.packages("tidyr")
}
if (!requireNamespace("zoo", quietly = TRUE)) {
install.packages("zoo")
}
if (!requireNamespace("patchwork", quietly = TRUE)) {
install.packages("patchwork")
}
# load packages
library("survival")
library("survminer")
## Loading required package: ggplot2
## Loading required package: ggpubr
##
## Attaching package: 'survminer'
## The following object is masked from 'package:survival':
##
## myeloma
library("ggplot2")
library("dplyr")
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
library("tidyr")
library("zoo")
##
## Attaching package: 'zoo'
## The following objects are masked from 'package:base':
##
## as.Date, as.Date.numeric
library("patchwork")KM 曲线
示例
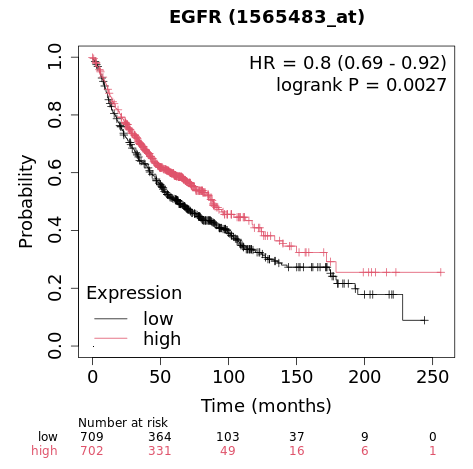
生存分析研究的是某个事件发生之前过去的时间,在临床研究中最常见的应用就是死亡率的估计(预测患者的生存时间),不过生存分析也可以应用于其他领域如机械故障时间等。
环境配置
系统要求: ## 跨平台(Linux/MacOS/Windows)
编程语言: R
依赖包:
survival,survminer,ggplot2,dplyr,tidyr,zoo, andpatchwork
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-04
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
dplyr * 1.1.4 2023-11-17 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
ggpubr * 0.6.2 2025-10-17 [1] RSPM
patchwork * 1.3.2 2025-08-25 [1] RSPM
survival * 3.8-3 2024-12-17 [3] CRAN (R 4.5.2)
survminer * 0.5.1 2025-09-02 [1] RSPM
tidyr * 1.3.2 2025-12-19 [1] RSPM
zoo * 1.8-15 2025-12-15 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
# 使用内置lung数据集(来自survival包)
data("lung", package = "survival")
# 数据预处理
surv_data <- lung %>%
mutate(
status = ifelse(status == 2, 1, 0), # 转换状态编码(1=事件)
sex = factor(sex, labels = c("Male", "Female")),
group = sample(c("Treatment", "Placebo"), n(), replace = TRUE)
)
# 查看数据结构
glimpse(surv_data)Rows: 228
Columns: 11
$ inst <dbl> 3, 3, 3, 5, 1, 12, 7, 11, 1, 7, 6, 16, 11, 21, 12, 1, 22, 16…
$ time <dbl> 306, 455, 1010, 210, 883, 1022, 310, 361, 218, 166, 170, 654…
$ status <dbl> 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ age <dbl> 74, 68, 56, 57, 60, 74, 68, 71, 53, 61, 57, 68, 68, 60, 57, …
$ sex <fct> Male, Male, Male, Male, Male, Male, Female, Female, Male, Ma…
$ ph.ecog <dbl> 1, 0, 0, 1, 0, 1, 2, 2, 1, 2, 1, 2, 1, NA, 1, 1, 1, 2, 2, 1,…
$ ph.karno <dbl> 90, 90, 90, 90, 100, 50, 70, 60, 70, 70, 80, 70, 90, 60, 80,…
$ pat.karno <dbl> 100, 90, 90, 60, 90, 80, 60, 80, 80, 70, 80, 70, 90, 70, 70,…
$ meal.cal <dbl> 1175, 1225, NA, 1150, NA, 513, 384, 538, 825, 271, 1025, NA,…
$ wt.loss <dbl> NA, 15, 15, 11, 0, 0, 10, 1, 16, 34, 27, 23, 5, 32, 60, 15, …
$ group <chr> "Placebo", "Placebo", "Placebo", "Treatment", "Placebo", "Pl…# 生存时间分布
summary(surv_data$time) Min. 1st Qu. Median Mean 3rd Qu. Max.
5.0 166.8 255.5 305.2 396.5 1022.0 # 拟合生存曲线
fit <- survfit(Surv(time, status) ~ group, data = surv_data)
# 提取曲线数据
surv_curve <- surv_summary(fit)
# 计算log-rank检验P值
diff <- survdiff(Surv(time, status) ~ group, data = surv_data)
p_value <- signif(1 - pchisq(diff$chisq, length(diff$n)-1), 3)可视化
1. 基础绘图
使用基础函数绘制图片的图注和简介。
# 基础生存曲线
p1 <- ggplot(surv_curve, aes(x = time, y = surv, color = strata)) +
geom_step(linewidth = 1) +
labs(x = "Time (Months)", y = "Survival Probability") +
scale_y_continuous(labels = scales::percent)
p1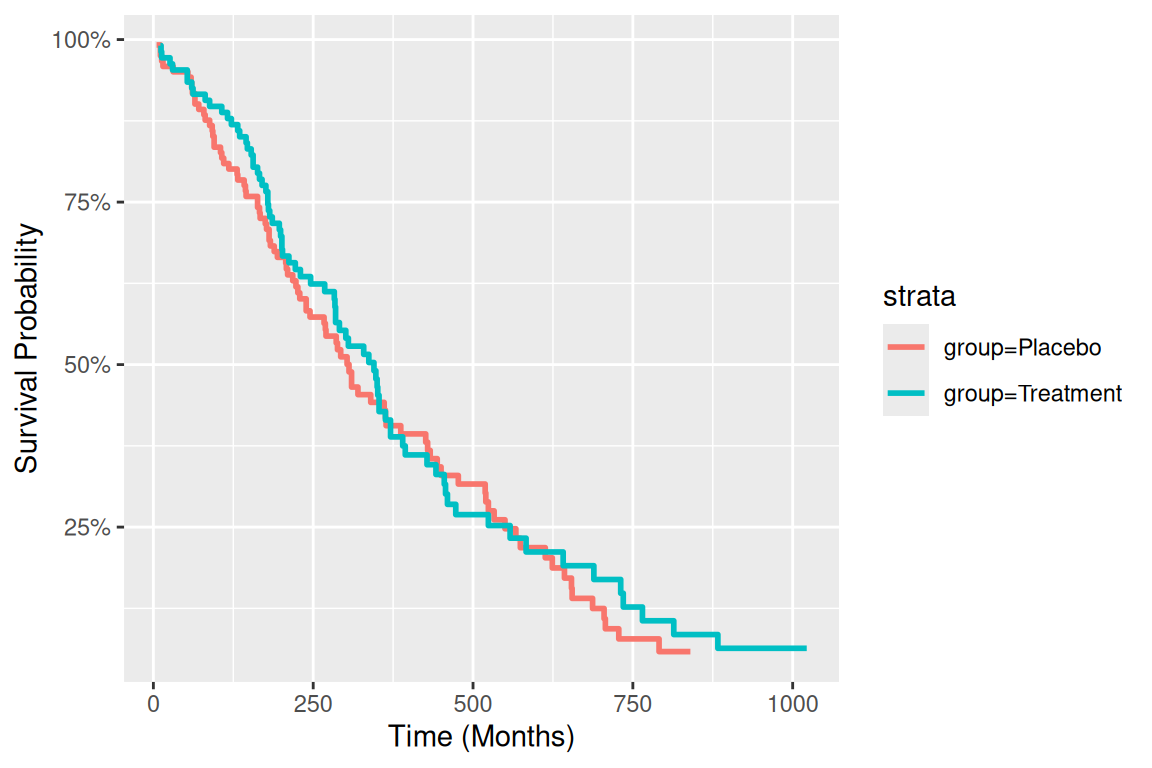
# 代码示例(以补充参数bins为例)-----
## 自定义美学映射
group_colors <- c("Treatment" = "#E64B35", "Placebo" = "#3182BD")
line_types <- c("Treatment" = "solid", "Placebo" = "dashed")
# 完整绘图代码
p_km <- ggplot(surv_curve, aes(x = time, color = group, fill = group)) +
# 先绘制置信区间
geom_ribbon(aes(ymin = lower, ymax = upper),
alpha = 0.15,
colour = NA) +
# 再绘制生存曲线
geom_step(aes(y = surv, linetype = group),
linewidth = 1.2) +
# 颜色映射
scale_color_manual(
values = group_colors,
name = "Treatment Group",
labels = c("Placebo", "Treatment")
) +
# 填充色映射
scale_fill_manual(
values = group_colors,
guide = "none"
) +
# 线型映射
scale_linetype_manual(
values = line_types,
guide = "none" # 与颜色共用图例
) +
# 坐标轴优化
scale_x_continuous(
name = "Time (Months)",
expand = c(0, 0),
breaks = seq(0, 1000, by = 100)
) +
scale_y_continuous(
name = "Survival Probability",
labels = scales::percent,
limits = c(0, 1)
) +
# 主题设置
theme_classic(base_size = 12) +
theme(
legend.position = c(0.85, 0.85),
legend.background = element_rect(fill = "white", colour = "grey50"),
panel.grid.major.y = element_line(colour = "grey90")
)
p_km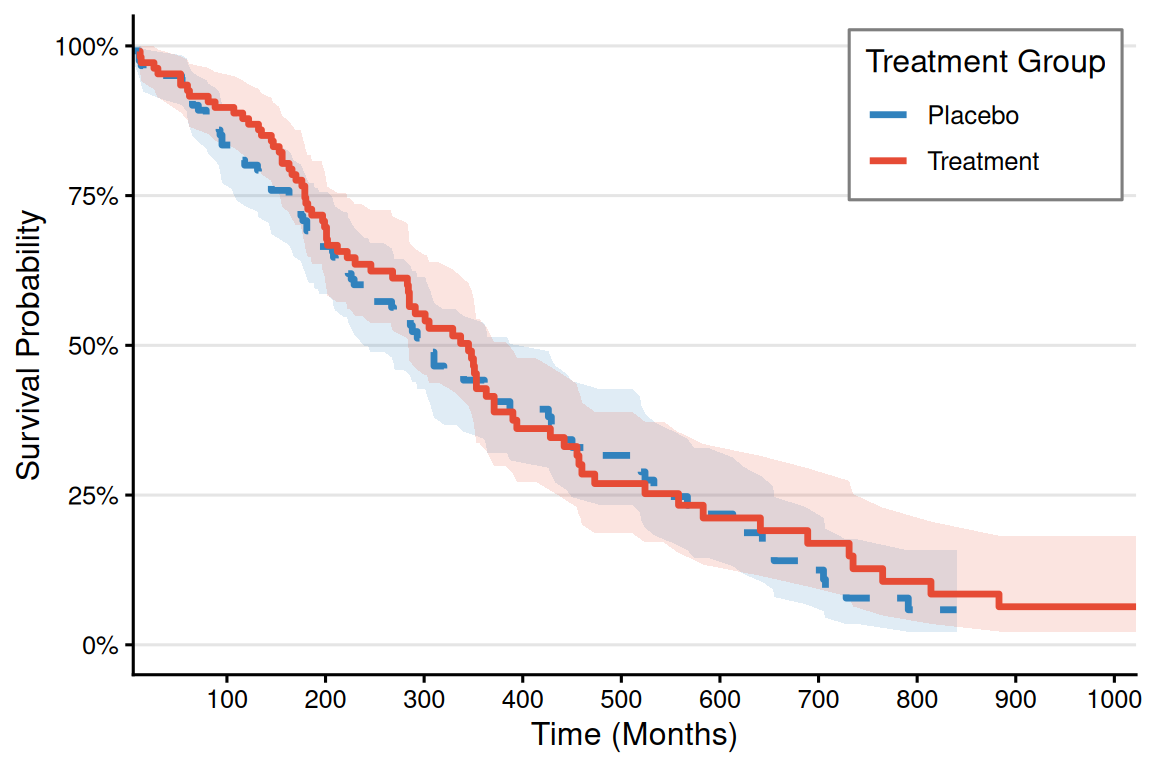
2. 更多进阶图表
# ----------------------------
# 风险表数据生成
# ----------------------------
# 生成分位数断点(自动适应数据分布)
custom_breaks <- quantile(surv_curve$time,
probs = seq(0, 1, 0.25),
na.rm = TRUE) %>%
unique() # 确保断点唯一
# 正确生成区间标签的方法
time_labels <- paste0(
round(custom_breaks[-length(custom_breaks)]), # 起始点(排除最后一个)
"-",
round(custom_breaks[-1]) # 结束点(排除第一个)
)
# 生成分段式风险表数据 --------------------------------------------------------
risk_table_segmented <- surv_curve %>%
mutate(
time_interval = cut(time,
breaks = custom_breaks,
include.lowest = TRUE,
right = FALSE,
labels = time_labels)
) %>%
group_by(group, time_interval) %>%
summarise(
n_risk = ifelse(all(is.na(n.risk)), 0, max(n.risk, na.rm = TRUE)), # 取区间内最大风险人数
.groups = "drop"
) %>%
complete(group, time_interval, fill = list(n_risk = 0)) %>%
# 4. 添加总计行(修复语法)
bind_rows(
group_by(., group) %>%
summarise(
time_interval = "Total",
n_risk = sum(n_risk, na.rm = TRUE),
.groups = "drop"
)
) %>%
mutate(
group = factor(group, levels = c("Treatment", "Placebo")),
time_interval = factor(time_interval, levels = c(time_labels, "Total") )
)
# 带边框的分段式风险表绘图 ----------------------------------------------------
risk_table_plot <- ggplot(risk_table_segmented,
aes(x = time_interval, y = group)) +
# # 单元格背景(带边框)
# geom_tile(color = "gray50", fill = "white", linewidth = 0.5,
# width = 0.95, height = 0.95) +
# 数值标签
geom_text(aes(label = n_risk), size = 4.5, fontface = "bold", color = "black") +
# 列标题样式
scale_x_discrete(
name = "Time Interval (Months)",
position = "top",
expand = expansion(add = 0.5)
) +
scale_y_discrete(
name = NULL,
limits = rev,
labels = c("Treatment\nGroup", "Placebo\nGroup") # 添加分组标签
) +
# 颜色系统
scale_fill_manual(values = group_colors, guide = "none") +
# 主题系统
theme_minimal(base_size = 12) +
theme(
axis.text.x = element_text(angle = 45, hjust = 0, color = "black"),
axis.text.y = element_text(
color = group_colors,
face = "bold",
margin = margin(r = 15)
),
panel.grid = element_blank(),
plot.margin = margin(t = 30, b = 5, unit = "pt"),
axis.title.x = element_text(margin = margin(t = 15))
) +
# 添加外边框
annotate("rect",
xmin = -Inf, xmax = Inf, ymin = -Inf, ymax = Inf,
color = "gray30", fill = NA, linewidth = 0.8)
risk_table_plot
# ----------------------------
# 绘图参数设置
# ----------------------------
group_colors <- c("Treatment" = "#E64B35", "Placebo" = "#3182BD")
theme_set(theme_minimal(base_size = 12))
# ----------------------------
# KM生存曲线绘制
# ----------------------------
p_km <- ggplot(surv_curve, aes(x = time, color = group, fill = group)) +
# 先绘制置信区间
geom_ribbon(aes(ymin = lower, ymax = upper),
alpha = 0.15,
colour = NA) +
# 再绘制生存曲线
geom_step(aes(y = surv, linetype = group),
linewidth = 1.2) +
# 颜色映射
scale_color_manual(
values = group_colors,
name = "Treatment Group",
labels = c("Placebo", "Treatment")
) +
# 填充色映射
scale_fill_manual(
values = group_colors,
guide = "none"
) +
# 线型映射
scale_linetype_manual(
values = line_types,
guide = "none" # 与颜色共用图例
) +
# 坐标轴优化
scale_x_continuous(
name = "Time (Months)",
expand = c(0, 0),
breaks = seq(0, 1000, by = 100)
) +
scale_y_continuous(
name = "Survival Probability",
labels = scales::percent,
limits = c(0, 1)
) +
# 主题设置
theme_classic(base_size = 12) +
theme(
legend.position = c(0.85, 0.85),
legend.background = element_rect(fill = "white", colour = "grey50"),
panel.grid.major.y = element_line(colour = "grey90")
)
p_km
# ----------------------------
# 图形组合
# ----------------------------
final_plot <- p_km / risk_table_plot +
plot_layout(heights = c(3, 0.4))
final_plot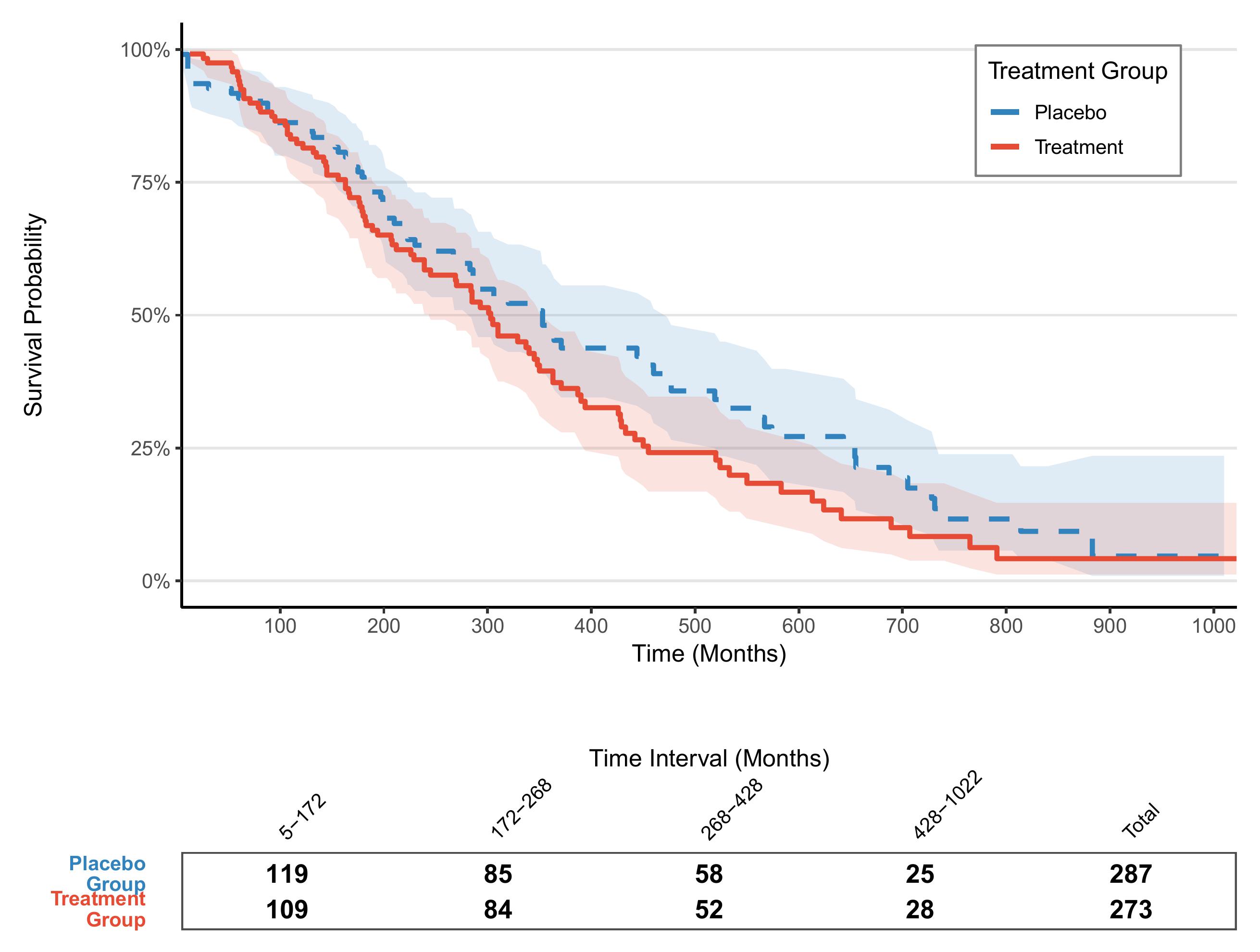
应用场景
1. 疾病分布
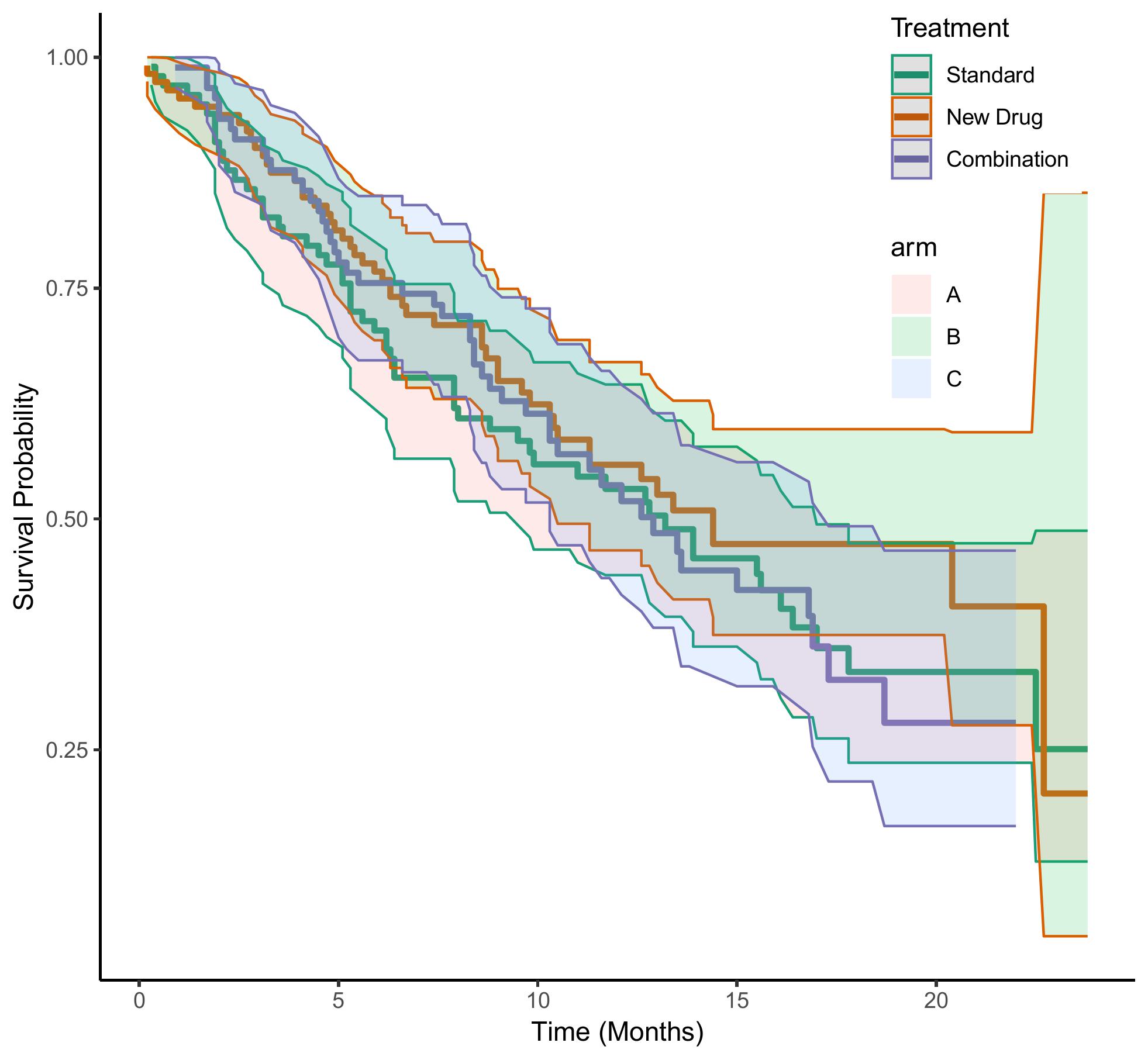
该图展示了 Standard、New Drug、Combination 三组数据之间的生成生存时间比较。
参考文献
[1] Kassambara A, et al. Survminer: Drawing Survival Curves using ggplot2. JOSS 2017
[2] Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer 2016
