# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("survminer", quietly = TRUE)) {
install.packages("survminer")
}
if (!requireNamespace("survival", quietly = TRUE)) {
install.packages("survival")
}
if (!requireNamespace("ggplotify", quietly = TRUE)) {
install.packages("ggplotify")
}
# 加载包
library(data.table)
library(jsonlite)
library(survminer)
library(survival)
library(ggplotify)生存分析
注记
Hiplot 网站
本页面为 Hiplot Survival Analysis 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
生存曲线是显示特定物种或群体 (如雄性或雌性) 在每个年龄存活下来的个体数量或比例的图表。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;survminer;survival;ggplotify
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-28
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.0 2025-12-24 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
ggplotify * 0.1.3 2025-09-20 [1] RSPM
ggpubr * 0.6.2 2025-10-17 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
survival * 3.8-3 2024-12-17 [3] CRAN (R 4.5.2)
survminer * 0.5.1 2025-09-02 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
载入数据为时间点,生存状态及分组。
# 加载数据
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/survival/data.json")$exampleData[[1]]$textarea[[1]])
data <- as.data.frame(data)
# 整理数据格式
colnames(data) <- c("Time", "Status", "Group")
data[,1] <- as.numeric(data[,1])
fit <- survfit(Surv(Time, Status == 1) ~ Group, data = data)
data <- data[data[,1] < 1100,]
# 查看数据
head(data) Time Status Group
1 306 1 G1
2 455 1 G1
3 1010 0 G1
4 210 1 G1
5 883 1 G1
6 1022 0 G1可视化
# 生存分析
p <- ggsurvplot(
fit, data = data, risk.table = T, pval = T, conf.int = T, fun = "pct",
size = 0.5, xlab = "Time", ylab = "Survival probability",
ggtheme = theme_bw(), risk.table.y.text.col = TRUE,
risk.table.height = 0.25, risk.table.y.text = T,
ncensor.plot = T, ncensor.plot.height = 0.25,
conf.int.style = "ribbon", surv.median.line = "hv",
palette = c("#00468BFF", "#ED0000FF"),
xlim = c(0, 1100), ylim = c(0, 100),
break.x.by = 150)
p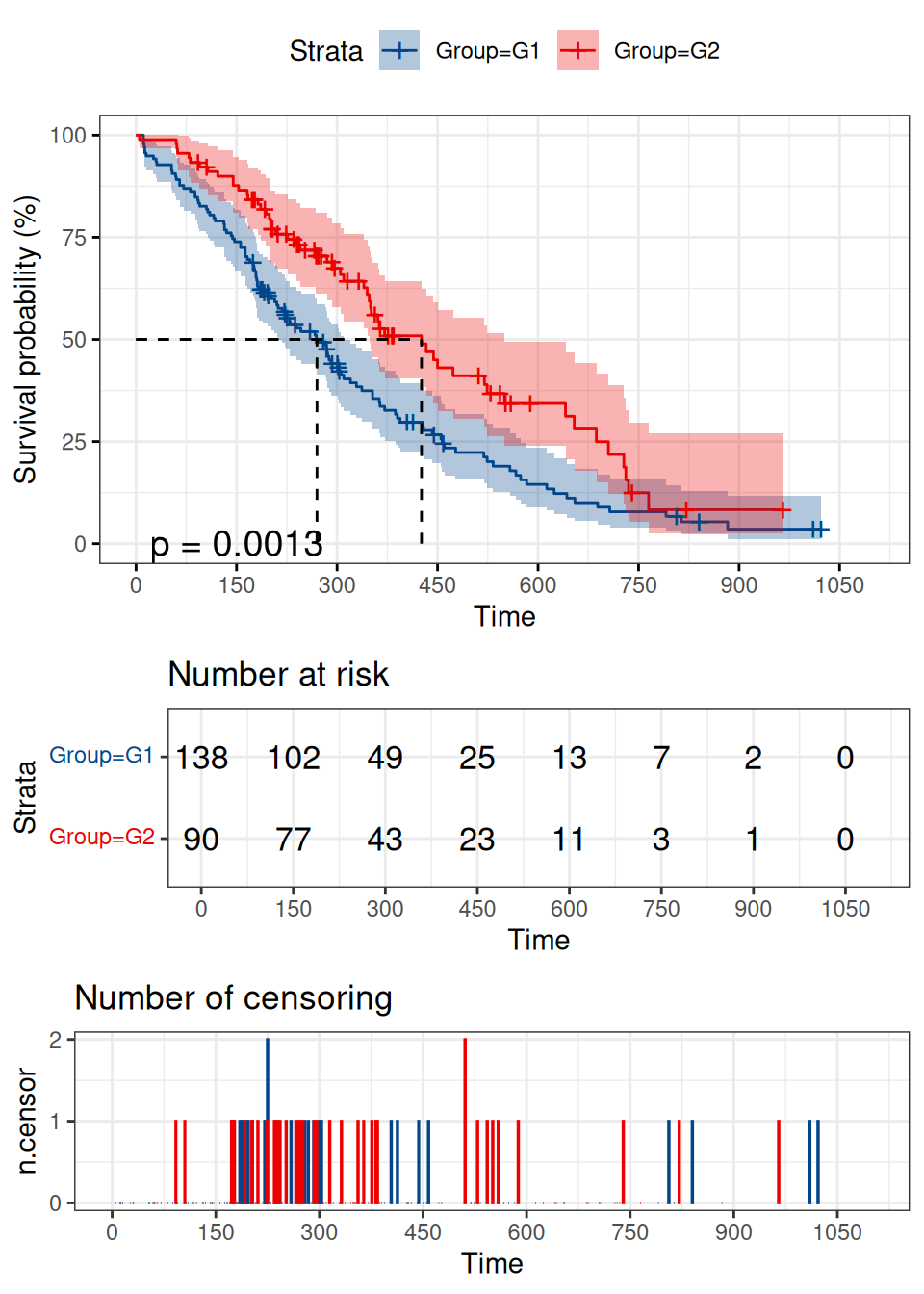
横轴表示时间轴,纵轴表示生存概率。蓝色曲线表示 G1 组生存曲线,红色曲线表示 G2 组生存曲线。经过 logrank 检验后发现 P 值 = 0.0013 < 0.05,表明两组患者生存状况的差异不能用抽样误差来解释,分组因素才是导致两条曲线生存率出现差异的原因。该示例图形表明,G2 组的总体生存率要好于 G1 组。
