# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("grafify", quietly = TRUE)) {
install.packages("grafify")
}
# 加载包
library(data.table)
library(jsonlite)
library(grafify)QQ 图
注记
Hiplot 网站
本页面为 Hiplot QQ Plot 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
用于直观验证一组数据是否来自某个分布,或者验证某两组数据是否来自同一(族)分布。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;grafify
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-28
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.0 2025-12-24 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
grafify * 5.1.0 2025-08-25 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
# 加载数据
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/qqplot/data.json")$exampleData[[1]]$textarea[[1]])
data <- as.data.frame(data)
# 整理数据格式
data[, "Genotype"] <- factor(data[, "Genotype"], levels = unique(data[, "Genotype"]))
# 查看数据
head(data) Genotype Cytokine Experiment facet
1 WT 1.312322 Exp_ 1 A
2 WT 2.171466 Exp_ 2 A
3 WT 1.497610 Exp_ 3 A
4 WT 1.088861 Exp_ 4 A
5 WT 1.270528 Exp_ 5 A
6 WT 3.012708 Exp_ 6 A可视化
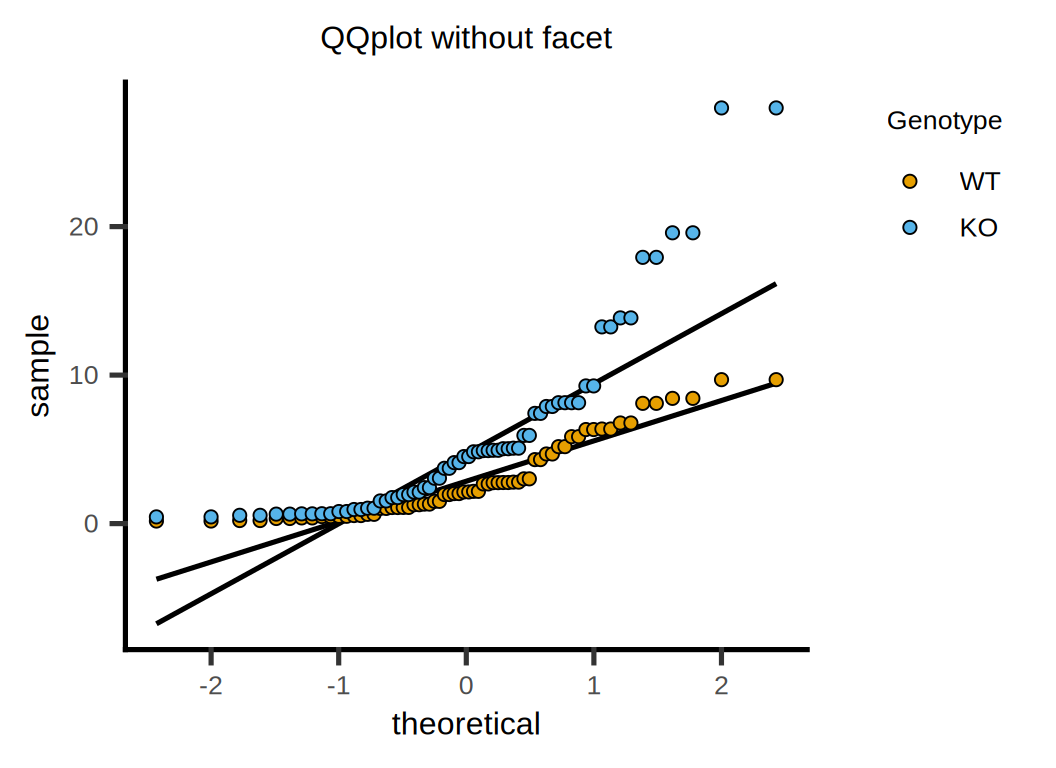
# QQ 图
p <- plot_qqline(data = data, ycol = Cytokine, group = Genotype,
symsize = 2, symthick = 0.5, s_alpha = 1) +
ggtitle("QQplot without facet") +
xlab("theoretical") + ylab("sample") +
guides(fill = guide_legend(title = "Genotype")) +
scale_color_manual(values = c("#E69F00","#4DB1DC")) +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p