# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("circlize", quietly = TRUE)) {
install.packages("circlize")
}
if (!requireNamespace("ComplexHeatmap", quietly = TRUE)) {
BiocManager::install("ComplexHeatmap")
}
if (!requireNamespace("gtrellis", quietly = TRUE)) {
remotes::install_github("jokergoo/gtrellis")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("ggplotify", quietly = TRUE)) {
install.packages("ggplotify")
}
if (!requireNamespace("RColorBrewer", quietly = TRUE)) {
install.packages("RColorBrewer")
}
# 加载包
library(data.table)
library(jsonlite)
library(circlize)
library(ComplexHeatmap)
library(gtrellis)
library(tidyverse)
library(ggplotify)
library(RColorBrewer)基因密度图
注记
Hiplot 网站
本页面为 Hiplot Gene Density 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
染色体数据展示。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;circlize;ComplexHeatmap;gtrellis;tidyverse;ggplotify;RColorBrewer
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-28
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
BiocGenerics * 0.56.0 2025-10-29 [1] Bioconduc~
circlize * 0.4.17 2025-12-08 [1] RSPM
ComplexHeatmap * 2.26.0 2025-10-29 [1] Bioconduc~
data.table * 1.18.0 2025-12-24 [1] RSPM
dplyr * 1.1.4 2023-11-17 [1] RSPM
forcats * 1.0.1 2025-09-25 [1] RSPM
generics * 0.1.4 2025-05-09 [1] RSPM
GenomicRanges * 1.62.1 2025-12-08 [1] Bioconduc~
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
ggplotify * 0.1.3 2025-09-20 [1] RSPM
gtrellis * 1.35.1 2025-11-02 [1] Github (jokergoo/gtrellis@86749f0)
IRanges * 2.44.0 2025-10-29 [1] Bioconduc~
jsonlite * 2.0.0 2025-03-27 [1] RSPM
lubridate * 1.9.4 2024-12-08 [1] RSPM
purrr * 1.2.1 2026-01-09 [1] RSPM
RColorBrewer * 1.1-3 2022-04-03 [1] RSPM
readr * 2.1.6 2025-11-14 [1] RSPM
S4Vectors * 0.48.0 2025-10-29 [1] Bioconduc~
Seqinfo * 1.0.0 2025-10-29 [1] Bioconduc~
stringr * 1.6.0 2025-11-04 [1] RSPM
tibble * 3.3.1 2026-01-11 [1] RSPM
tidyr * 1.3.2 2025-12-19 [1] RSPM
tidyverse * 2.0.0 2023-02-22 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
# 加载数据
data1 <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/gene-density/data.json")$exampleData$textarea[[1]])
data1 <- as.data.frame(data1)
data2 <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/gene-density/data.json")$exampleData$textarea[[2]])
data2 <- as.data.frame(data2)
# 整理数据格式
chrNum <- str_replace(unique(data1$chr), "Chr|chr", "")
data1$chr <- factor(data1$chr, levels = paste0("Chr", chrNum))
data2$chr <- factor(data2$chr, levels = paste0("Chr", chrNum))
# 设置窗口计算基因密度
windows <- 100 * 1000 # 默认:100kb
gene_density <- genomicDensity(data2, window.size = windows)
gene_density$chr <- factor(gene_density$chr,
levels = paste0("Chr", chrNum)
)
# 查看数据
head(data1) chr start end
1 Chr5 0 29958434
2 Chr8 0 28443022
3 Chr9 0 23012720
4 Chr10 0 23207287
5 Chr12 0 27531856head(data2) chr start end
1 Chr10 38648 40060
2 Chr10 45941 58338
3 Chr10 67119 72971
4 Chr10 75410 76305
5 Chr10 80964 82250
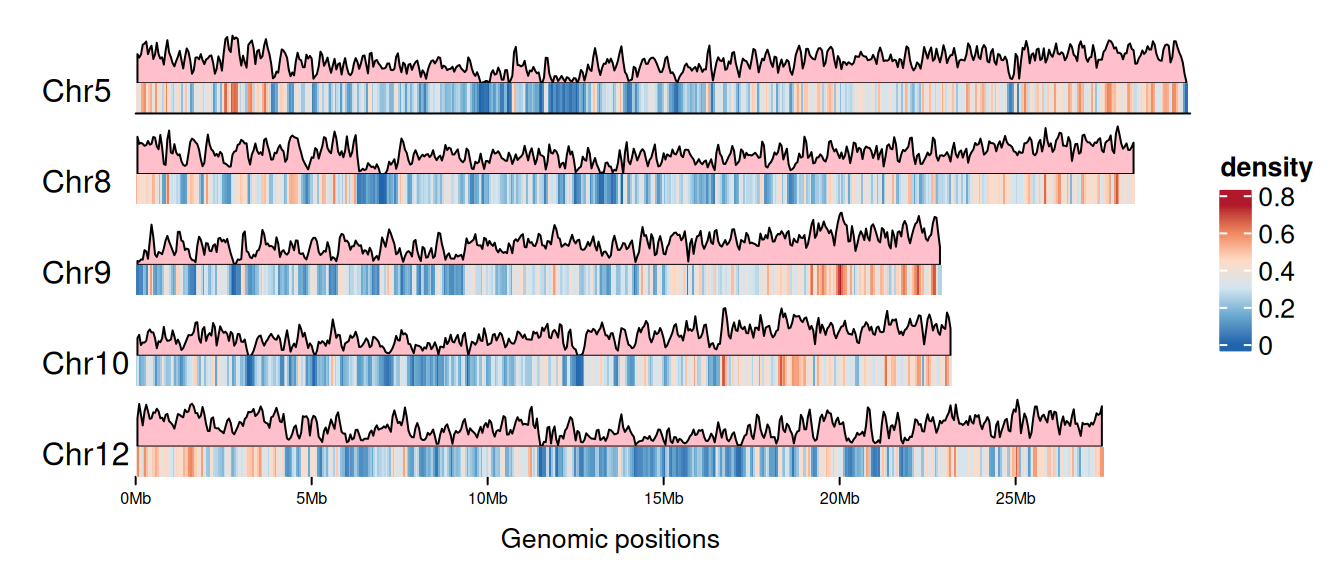
6 Chr10 94798 97746可视化
# 设置画板颜色
palettes <- c("#B2182B","#EF8A62","#FDDBC7","#D1E5F0","#67A9CF","#2166AC")
col_fun <- colorRamp2(
seq(0, max(gene_density[[4]]), length = 6), rev(palettes)
)
cm <- ColorMapping(col_fun = col_fun)
# 设置图例
lgd <- color_mapping_legend(
cm, plot = F, title = "density", color_bar = "continuous"
)
# 绘制基因密度分布热图
p <- as.ggplot(function() {
gtrellis_layout(
data1, n_track = 2, ncol = 1, byrow = FALSE,
track_axis = FALSE, add_name_track = FALSE,
xpadding = c(0.1, 0), gap = unit(1, "mm"),
track_height = unit.c(unit(1, "null"), unit(4, "mm")),
track_ylim = c(0, max(gene_density[[4]]), 0, 1),
border = FALSE, asist_ticks = FALSE,
legend = lgd
)
# 添加基因面积图 track
add_lines_track(gene_density, gene_density[[4]],
area = TRUE, gp = gpar(fill = "pink"))
# 添加基因密度热图 track
add_heatmap_track(gene_density, gene_density[[4]], fill = col_fun)
add_track(track = 2, clip = FALSE, panel_fun = function(gr) {
chr <- get_cell_meta_data("name")
if (chr == paste("Chr", length(chrNum), sep = "")) {
grid.lines(get_cell_meta_data("xlim"), unit(c(0, 0), "npc"),
default.units = "native")
}
grid.text(chr, x = 0.01, y = 0.38, just = c("left", "bottom"))
})
circos.clear()
})
p