# 安装包
if (!requireNamespace("ComplexHeatmap", quietly = TRUE)) {
BiocManager::install("ComplexHeatmap")
}
if (!requireNamespace("dendextend", quietly = TRUE)) {
install.packages("dendextend")
}
if (!requireNamespace("tidyr", quietly = TRUE)) {
install.packages("tidyr")
}
if (!requireNamespace("circlize", quietly = TRUE)) {
install.packages("circlize")
}
if (!requireNamespace("gridExtra", quietly = TRUE)) {
install.packages("gridExtra")
}
if (!requireNamespace("pheatmap", quietly = TRUE)) {
install.packages("pheatmap")
}
# 加载包
library(ComplexHeatmap)
library(dendextend)
library(tidyr)
library(circlize)
library(gridExtra)
library(pheatmap)复杂热图
示例
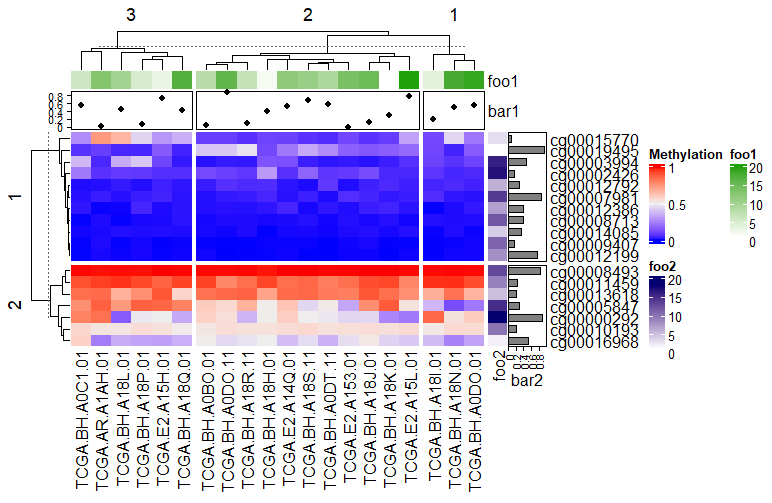
复杂热图(Complex Heatmap)是一种高级数据可视化工具,主要用于展示多维数据的矩阵关系,并在基础热图(Heatmap)上增加了多层次注释、聚类分析、数据拆分等功能。它能够更清晰地呈现复杂数据的内在结构和关联性。
环境配置
系统要求: 跨平台(Linux/MacOS/Windows)
编程语言:R
依赖包:
ComplexHeatmap,dendextend,tidyr,circlize,gridExtra,pheatmap
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-01
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
circlize * 0.4.17 2025-12-08 [1] RSPM
ComplexHeatmap * 2.26.0 2025-10-29 [1] Bioconduc~
dendextend * 1.19.1 2025-07-15 [1] RSPM
gridExtra * 2.3 2017-09-09 [1] RSPM
pheatmap * 1.0.13 2025-06-05 [1] RSPM
tidyr * 1.3.2 2025-12-19 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
数据主要来自TCGA数据库的甲基化数据
#data_mat(连续型)
data <- readr::read_csv("https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/TCGA.BRCA.sampleMap_HumanMethylation27_ch.csv")
data <- as.data.frame(data)
rownames(data) <- data[,1]
data <- data[,-1]
data_TCGA <- na.omit(data[1:20,1:20])
data_TCGA <- as.matrix(data_TCGA)
#离散型
discrete_mat = matrix(sample(1:5, 100, replace = TRUE), 10, 10)可视化
1. 基础复杂热图
连续型变量
这里我们以TCGA数据库的甲基化数据为例
# 连续型变量
Heatmap(data_TCGA , name = "Methylation")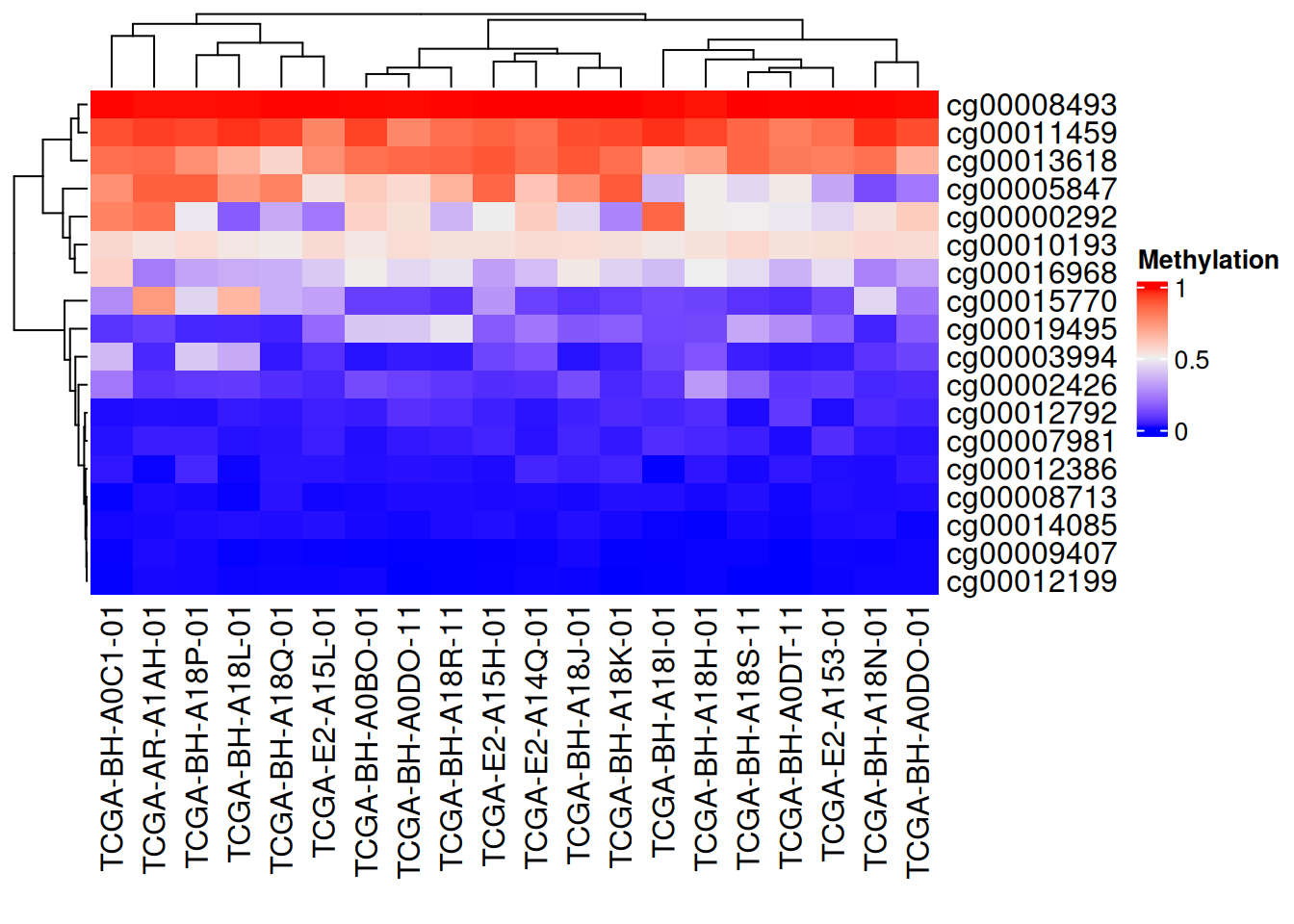
上图为甲基化水平的复杂热图,列为基因位点,行为样本。红色表示高甲基化水平,蓝色表示低甲基化水平。边轴为聚类分析结果。
离散型变量
这里我们以生成数据为例
# 离散型变量
Heatmap(discrete_mat, name = "mat")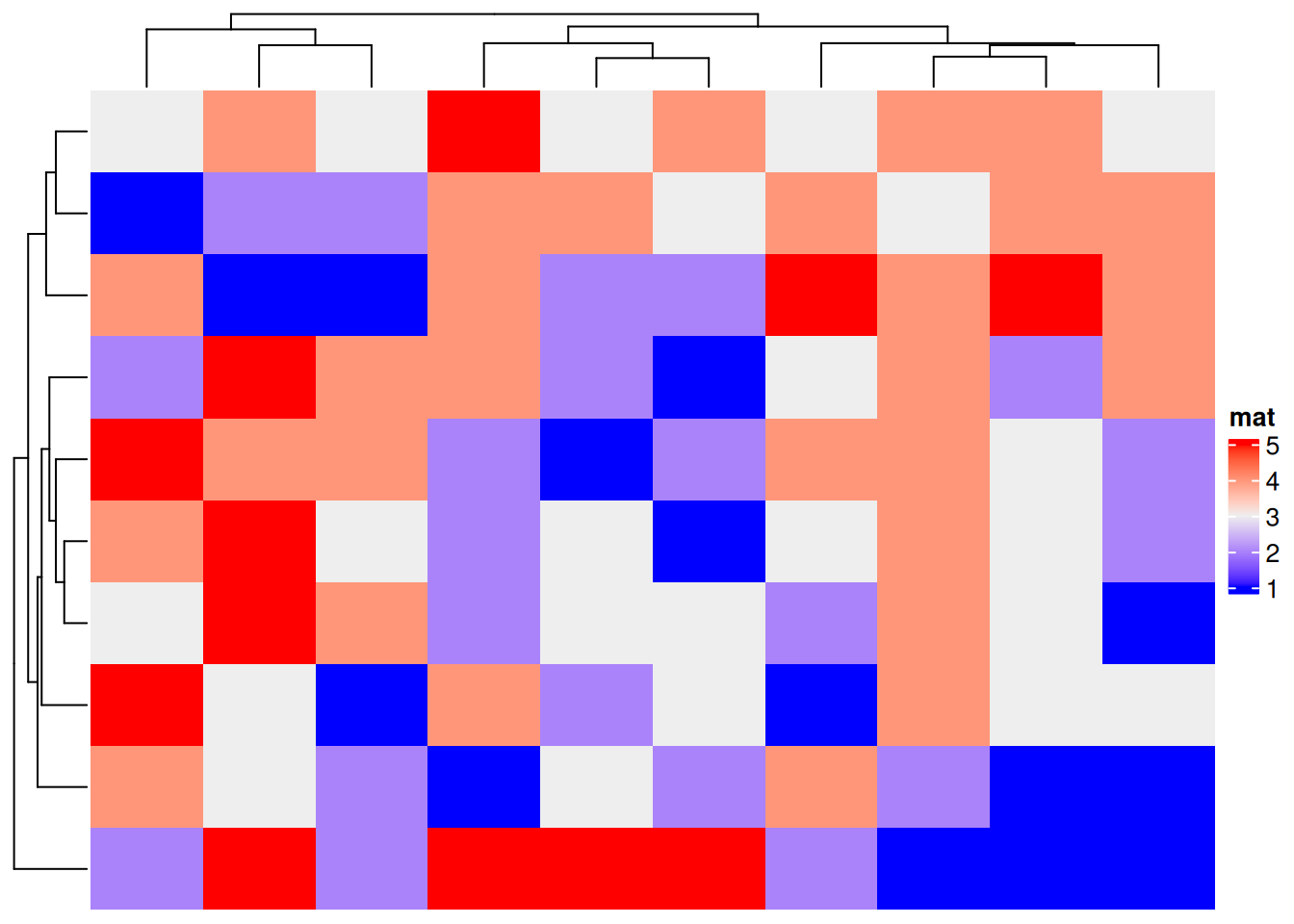
上图为生成的离散变量的复杂热图,边轴为聚类分析结果。
2. 个性化
2.1 颜色自定义
连续型变量
# 连续型变量颜色自定义
col_fun = colorRamp2(c(0, 0.5, 1), #设置截断点
c("blue", "white", "pink"))
Heatmap(data_TCGA, name = "Methylation", col = col_fun,
column_title = "自定义三段颜色")
# 连续型变量颜色自定义
Heatmap(data_TCGA, name = "Methylation", col = rev(rainbow(10)),
column_title = "创建颜色向量")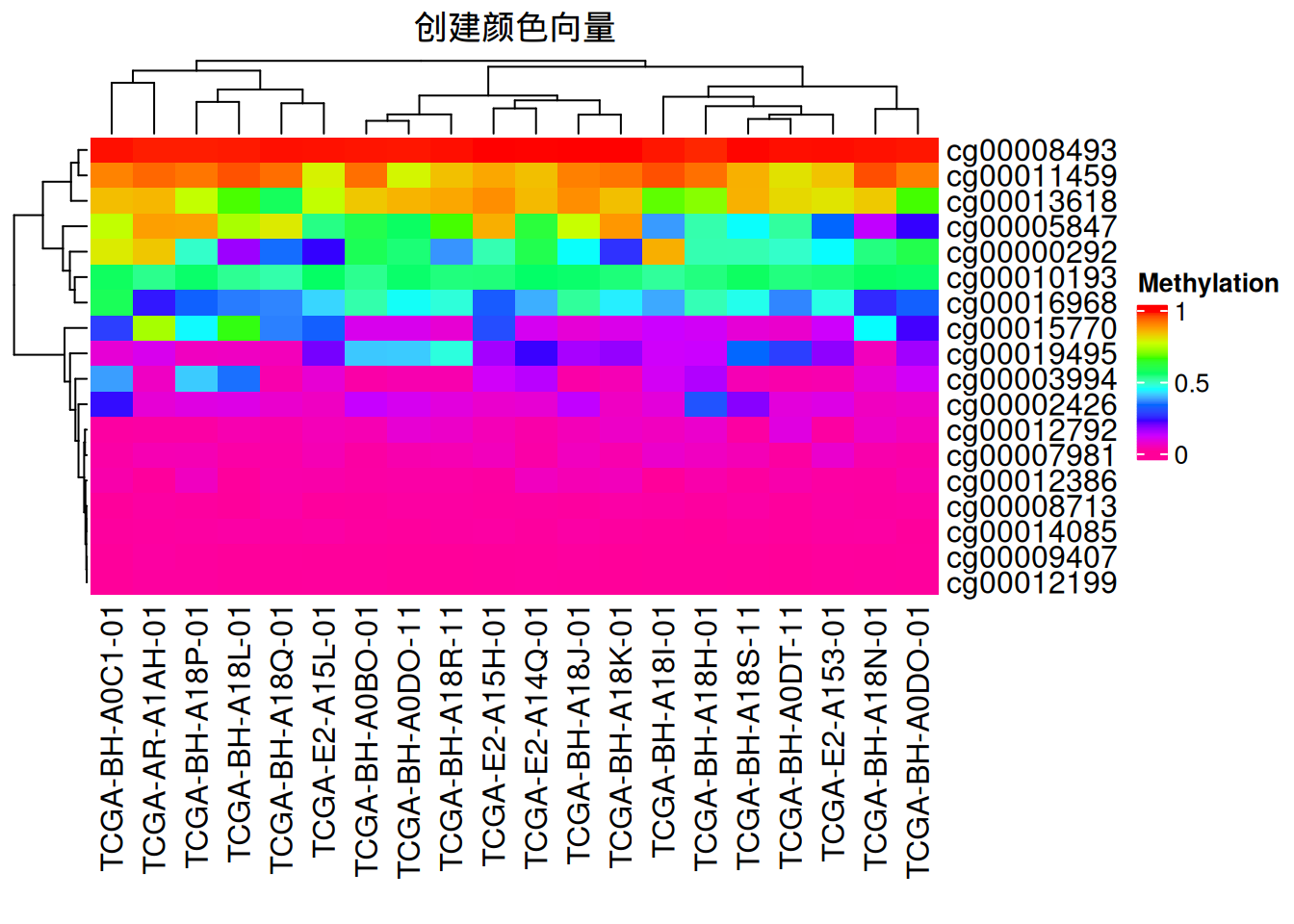
离散型变量
# 离散型变量颜色自定义
colors <- c(
"1" = "#BBEDA7",
"2" = "#F3D8FA",
"3" = "#FFECA1",
"4" = "#D9E6F7",
"5" = "#EFBEC6"
)
Heatmap(discrete_mat, name = "mat", col = colors,
column_title = "离散型变量颜色自定义")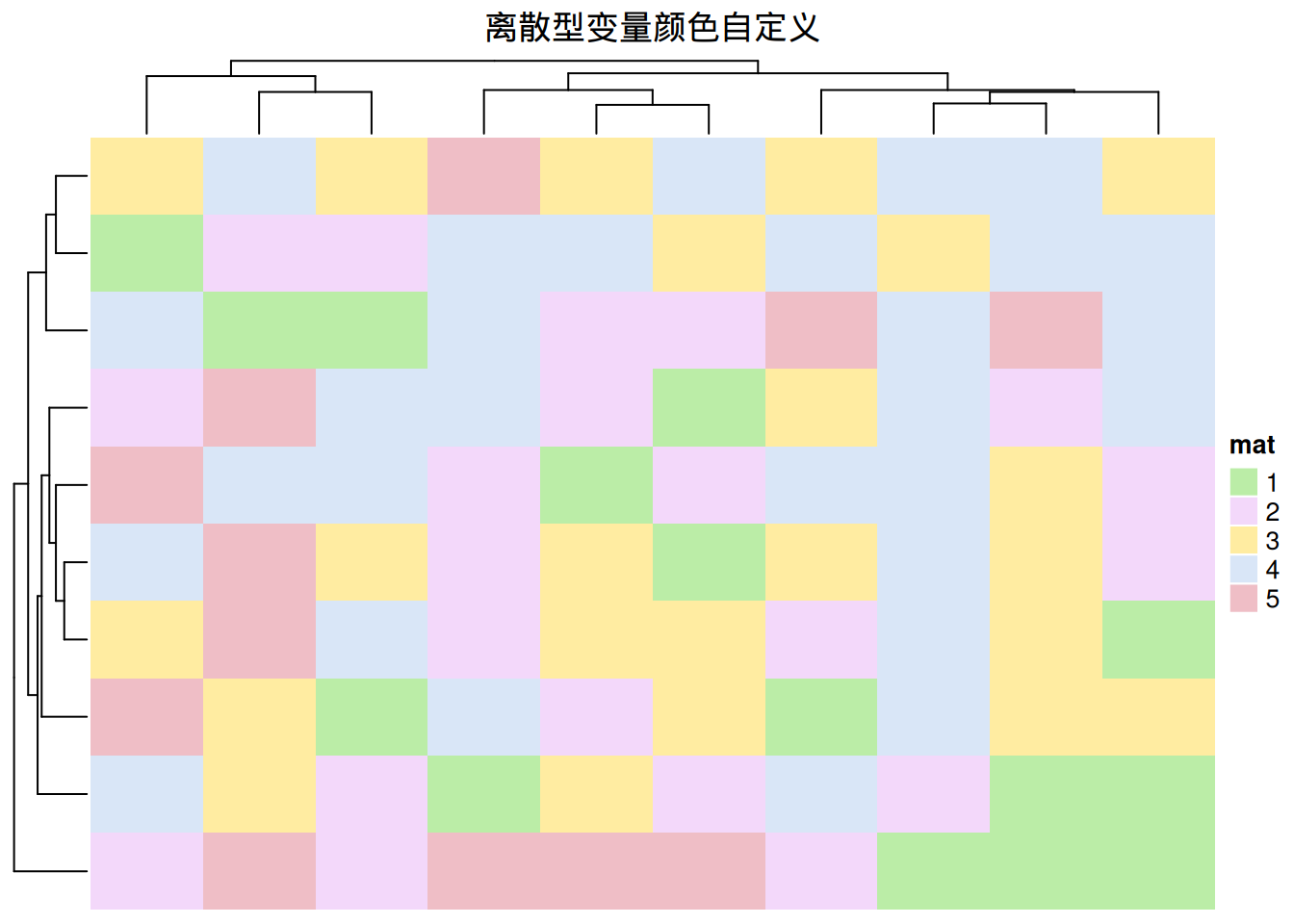
2.2 边框自定义
# 边框自定义
Heatmap(data_TCGA, name = "Methylation",
border = "black", #border值可为FALSE或者颜色
rect_gp = gpar(col = "white", lwd = 2),#lty线型, lwd线宽
column_title = "边框自定义")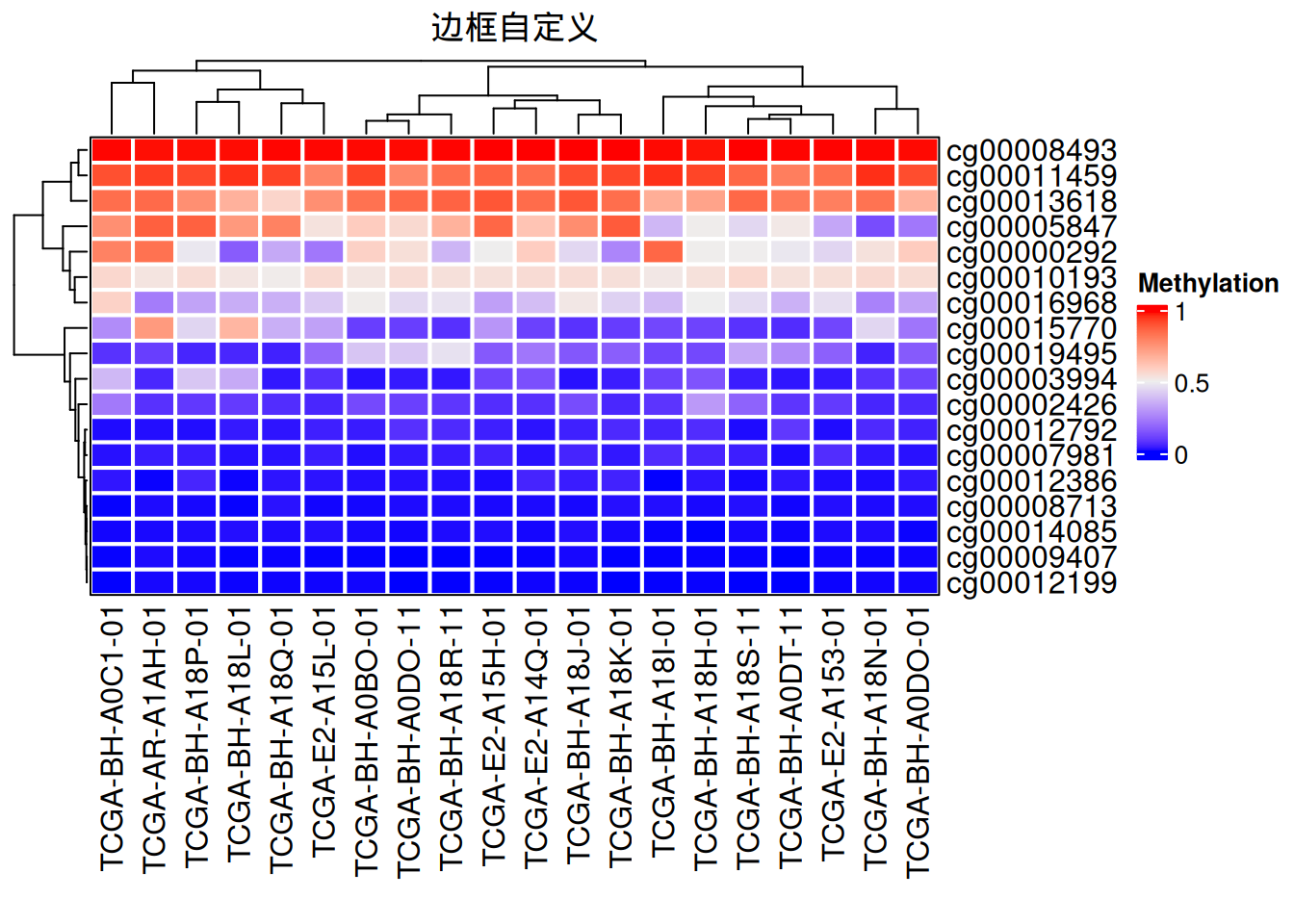
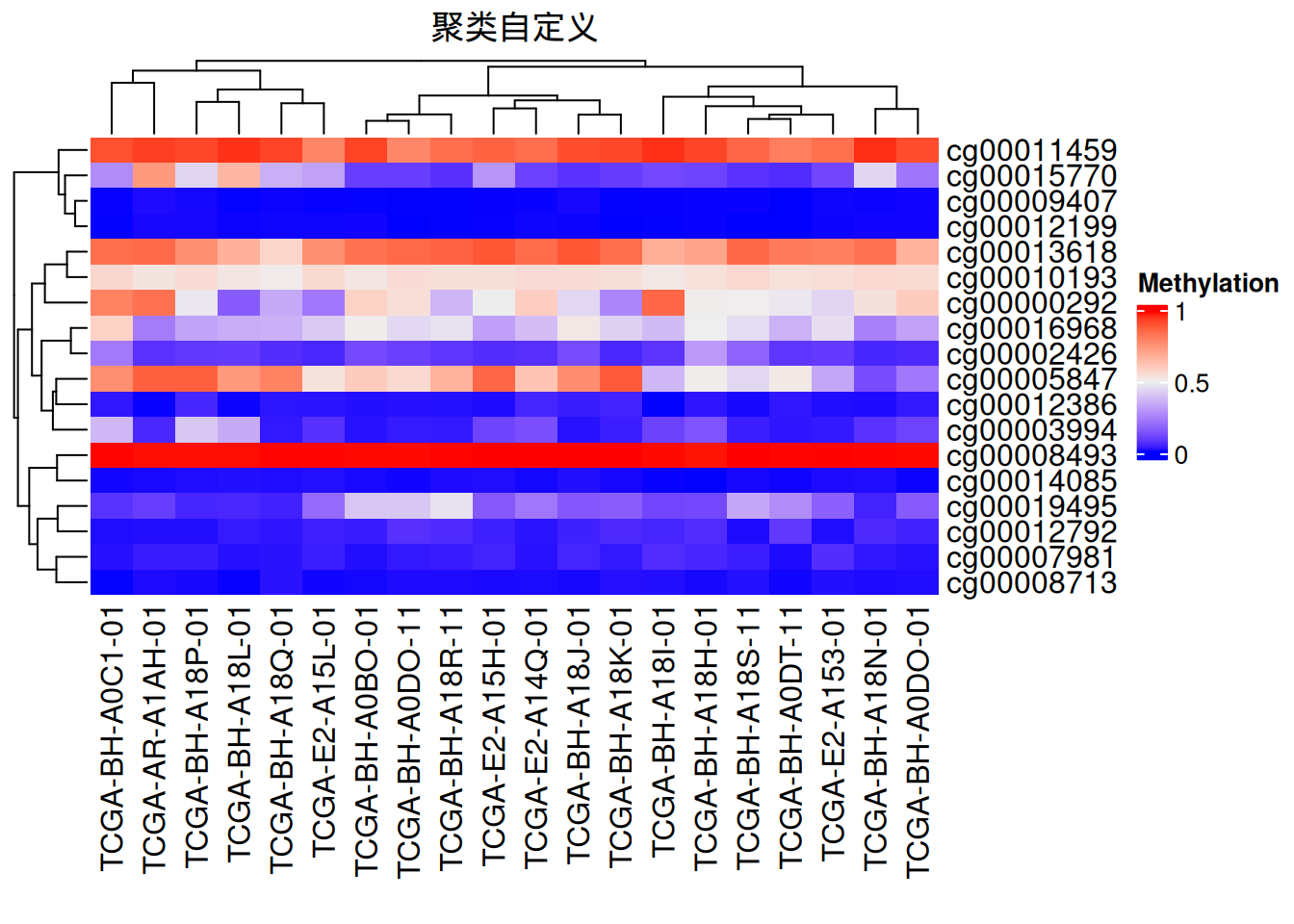
3. 聚类
可以自定义聚类方法,树状图位置、大小等
# 聚类
Heatmap(data_TCGA, name = "Methylation",
# row_dend_side = "right",
# column_dend_side = "bottom", #可调整树状图位置
column_dend_height = unit(1, "cm"),
row_dend_width = unit(1, "cm"),#可调整树状图距离
column_title = "聚类自定义",
clustering_distance_rows = "pearson", #聚类方法,可自设函数
# show_parent_dend_line = FALSE #可隐藏虚线
) 
树状图渲染
我们还利用dendextend包对树状图进行颜色渲染
# 树状图渲染
row_dend = as.dendrogram(hclust(dist(data_TCGA)))
row_dend = color_branches(row_dend, k = 2) #生成分支
#数字表示颜色数量
Heatmap(data_TCGA, name = "Methylation",
cluster_rows = row_dend,
# row_dend_gp = gpar(col = "red") #也可自定义颜色
)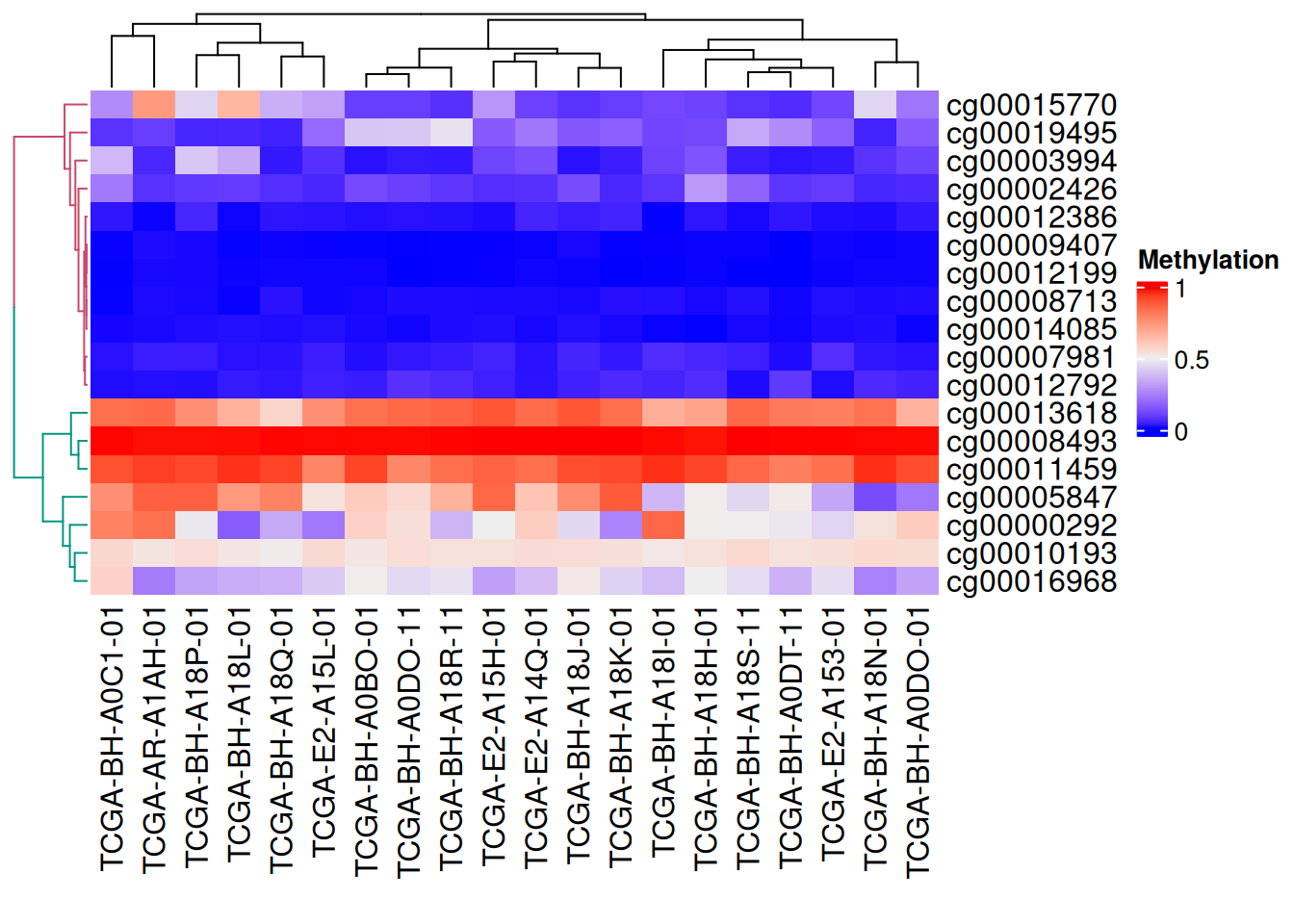
上图可见树状图分支为不同颜色
4. 分割
连续型变量
采用k-means聚类分割
# 连续型变量分割
Heatmap(data_TCGA,
name = "Methylation",
row_km = 2, #设置分割数
column_km = 3)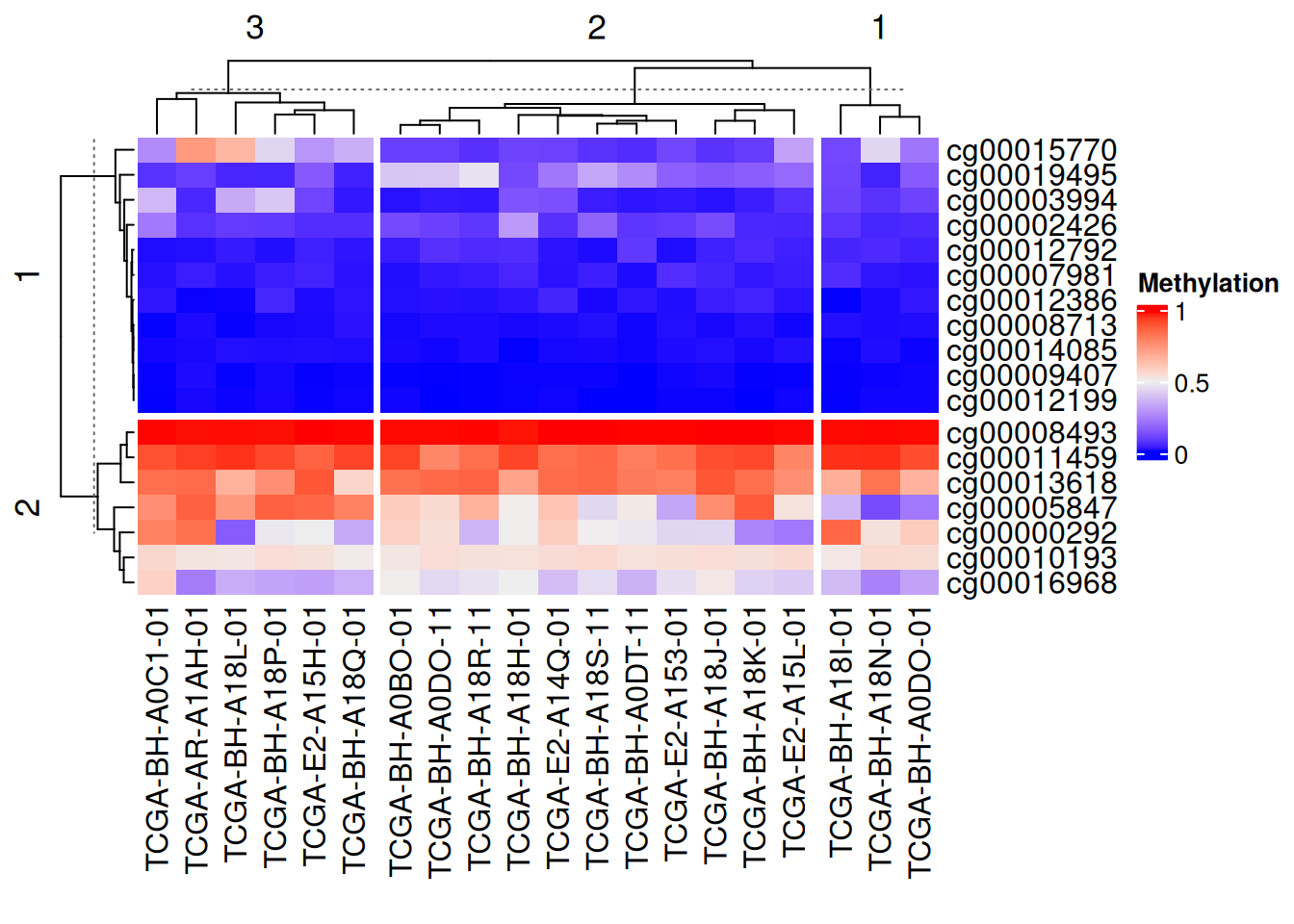
利用向量分割
# 利用向量分割
Heatmap(data_TCGA, name = "Methylation",
row_split = rep(c("A", "B"), 9),
column_split = rep(c("C", "D"), 10))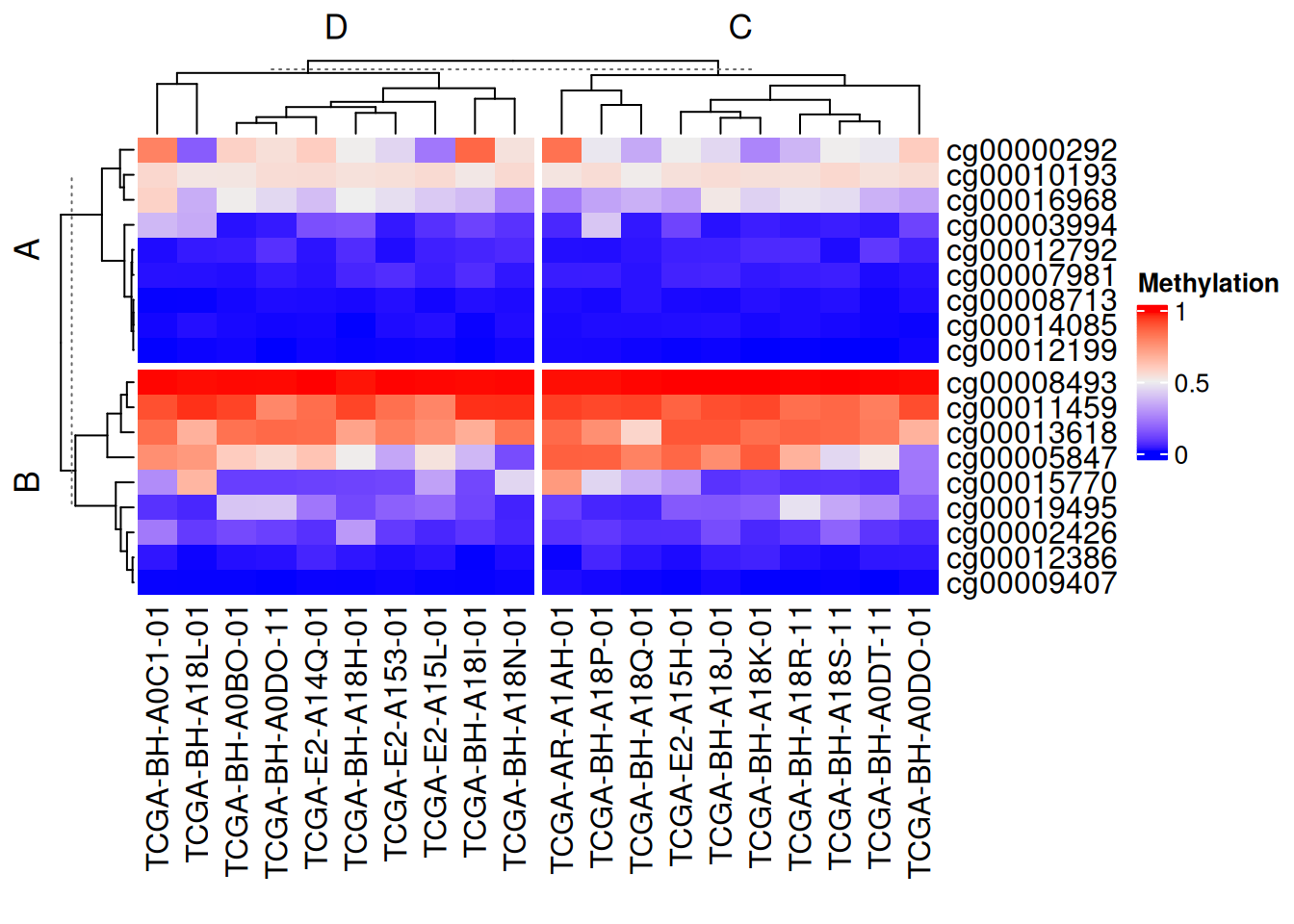
利用矩阵分割
# 利用矩阵分割
Heatmap(data_TCGA, name = "Methylation",
row_split = data.frame(rep(c("A", "B"), 9),
rep(c("C", "D"), each = 9)))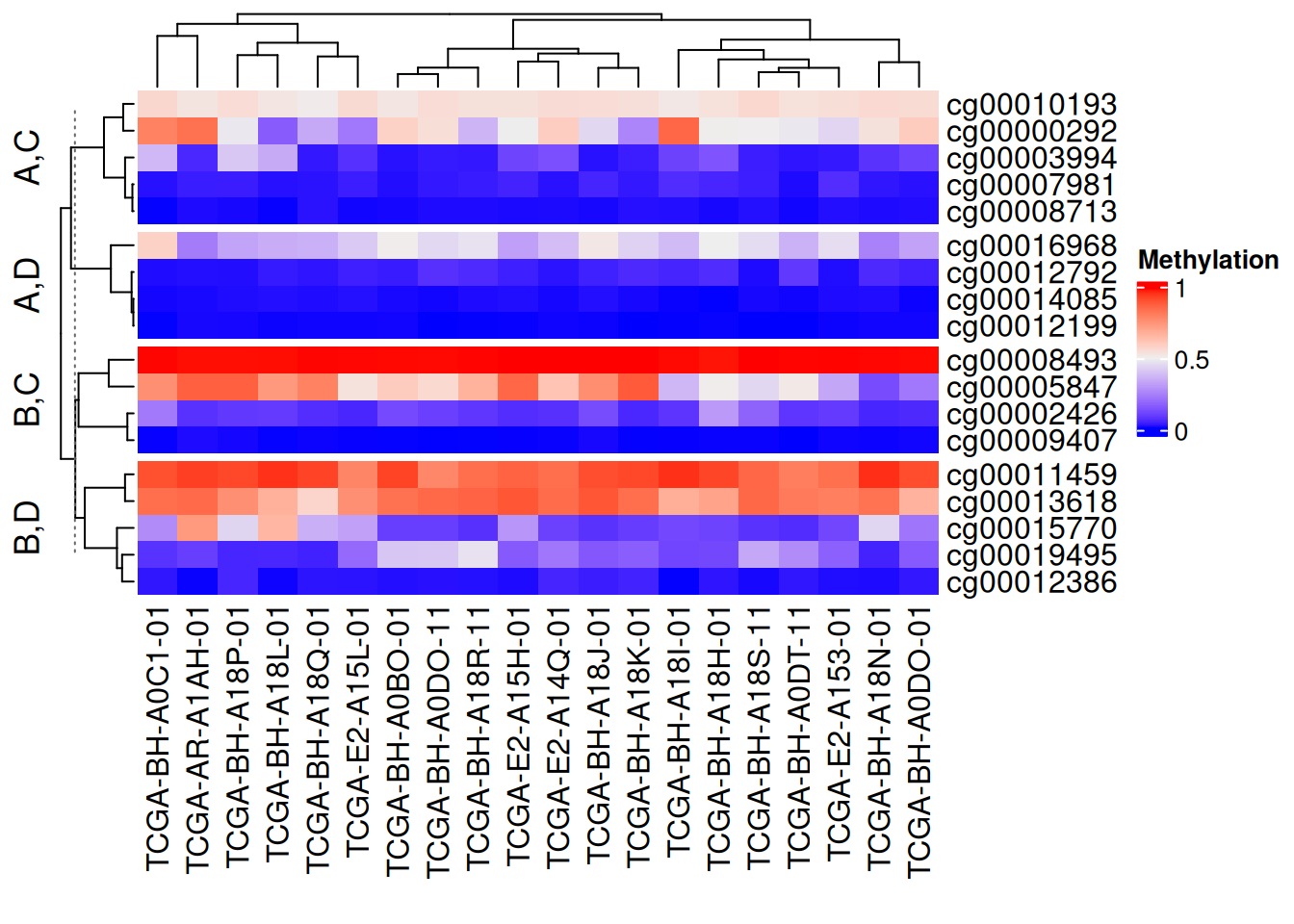
以上分割方法均可行列独立混使用,设置show_parent_dend_line = FALSE可隐藏虚线
离散型变量
按指定行列进行分割
# 按指定行列进行分割
Heatmap(discrete_mat,
name = "mat",
col = 1:4,
row_split = discrete_mat[,1])#根据矩阵数据中的第一列进行分割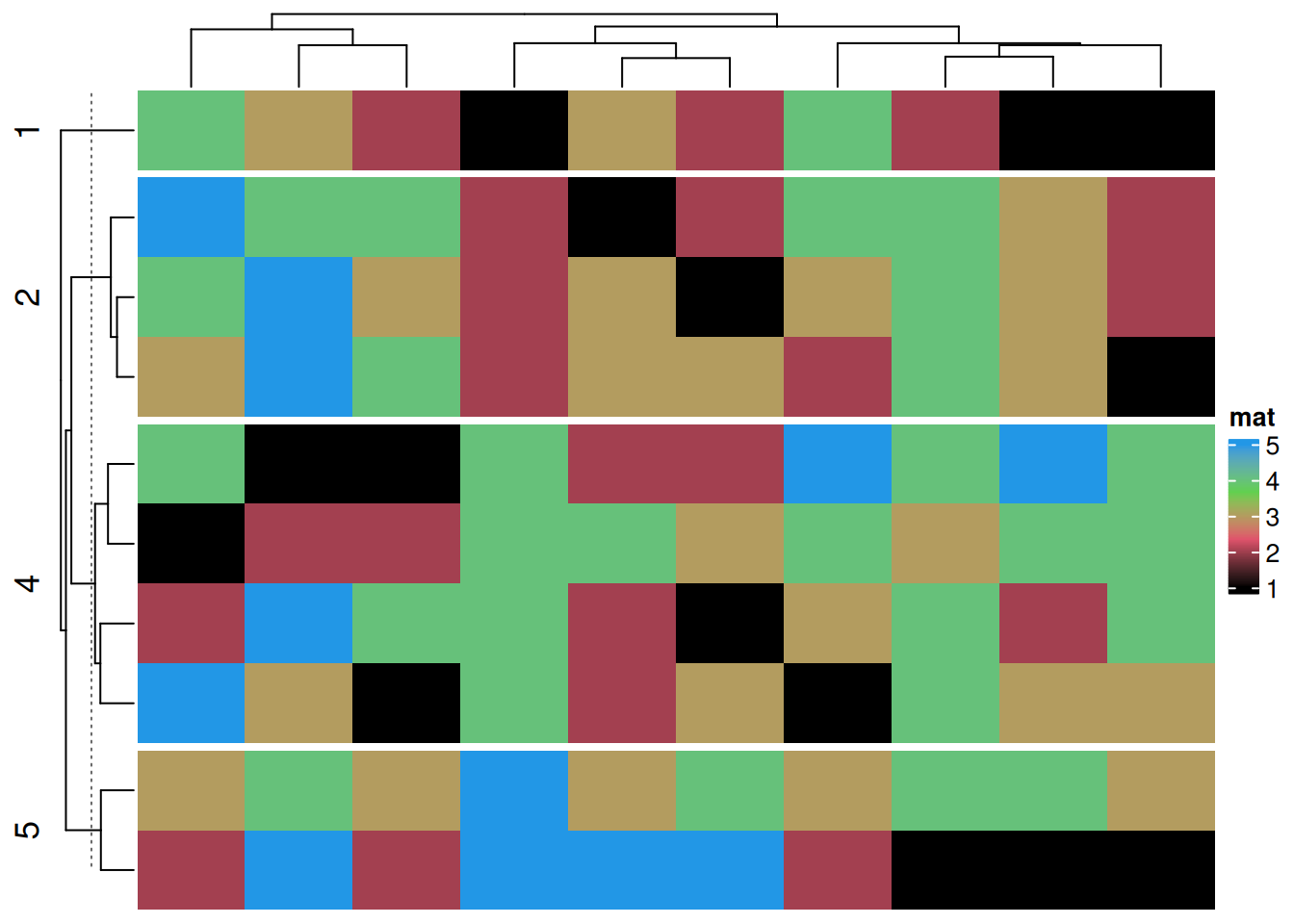
参数分割
可根据热图分割对标题,聚类参数等进行分割
# 参数分割
Heatmap(data_TCGA, name = "Methylation",
row_km = 2,
row_title_gp = gpar(col = c("#9A090A", "#0D4B99"), font = 1:2),
row_names_gp = gpar(col = c("#748901", "#E6AC03"), fontsize = c(10, 12)),
column_km = 3,
column_title_gp = gpar(fill = c("#BFD641", "#FE9900", "#CC6CE7"), font = 1:3), #fill用于填充
column_names_gp = gpar(col = c("#1D6103", "#E08804", "#5E0676"), fontsize = c(10, 12, 8)))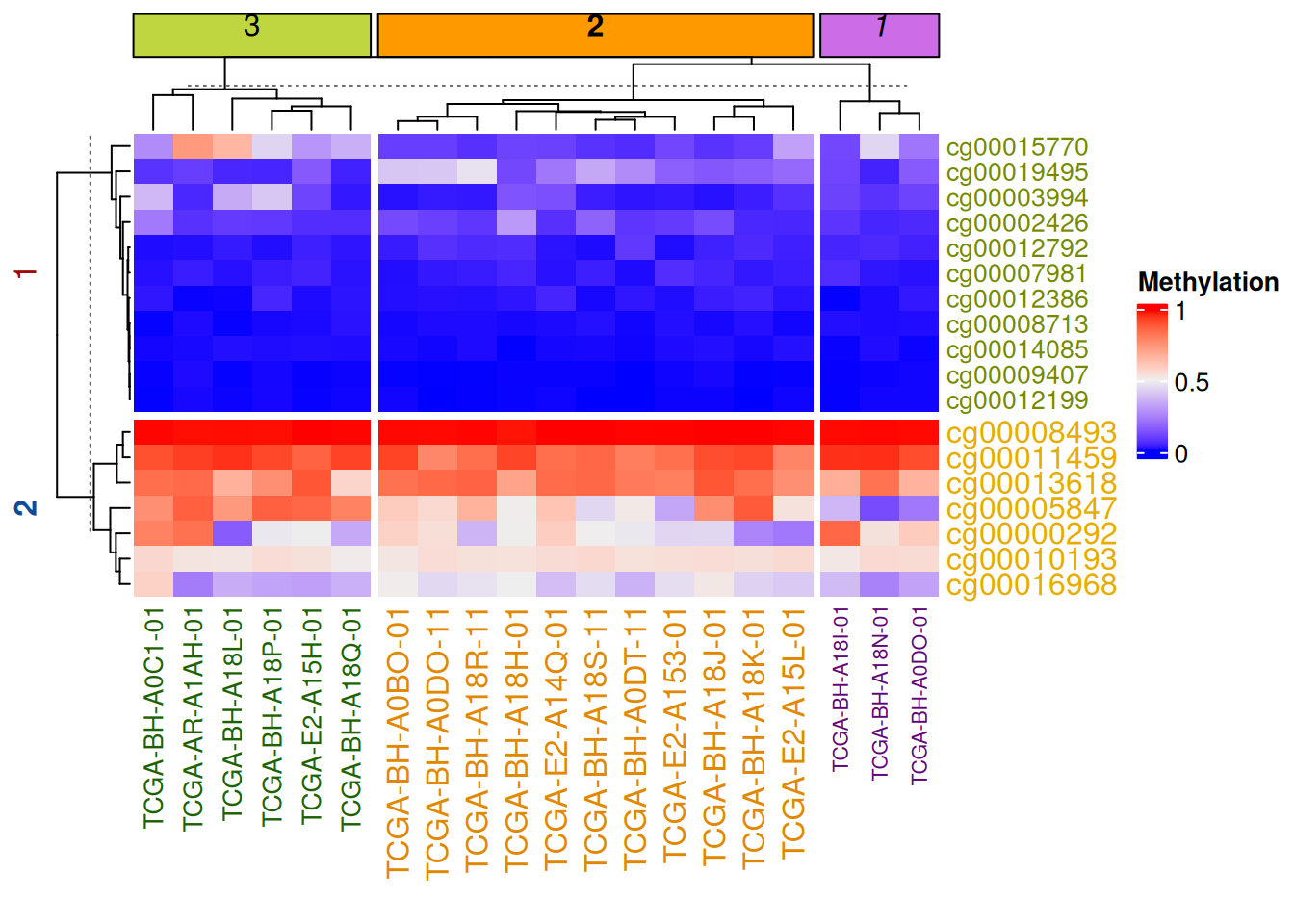
注释分割
可根据热图分割对边注释进行分割
# 注释分割
Heatmap(data_TCGA, name = "Methylation", row_km = 2, column_km = 3,
top_annotation = HeatmapAnnotation(foo1 = 1:20, bar1 = anno_points(runif(20))),
right_annotation = rowAnnotation(foo2 = 18:1, bar2 = anno_barplot(runif(18)))
)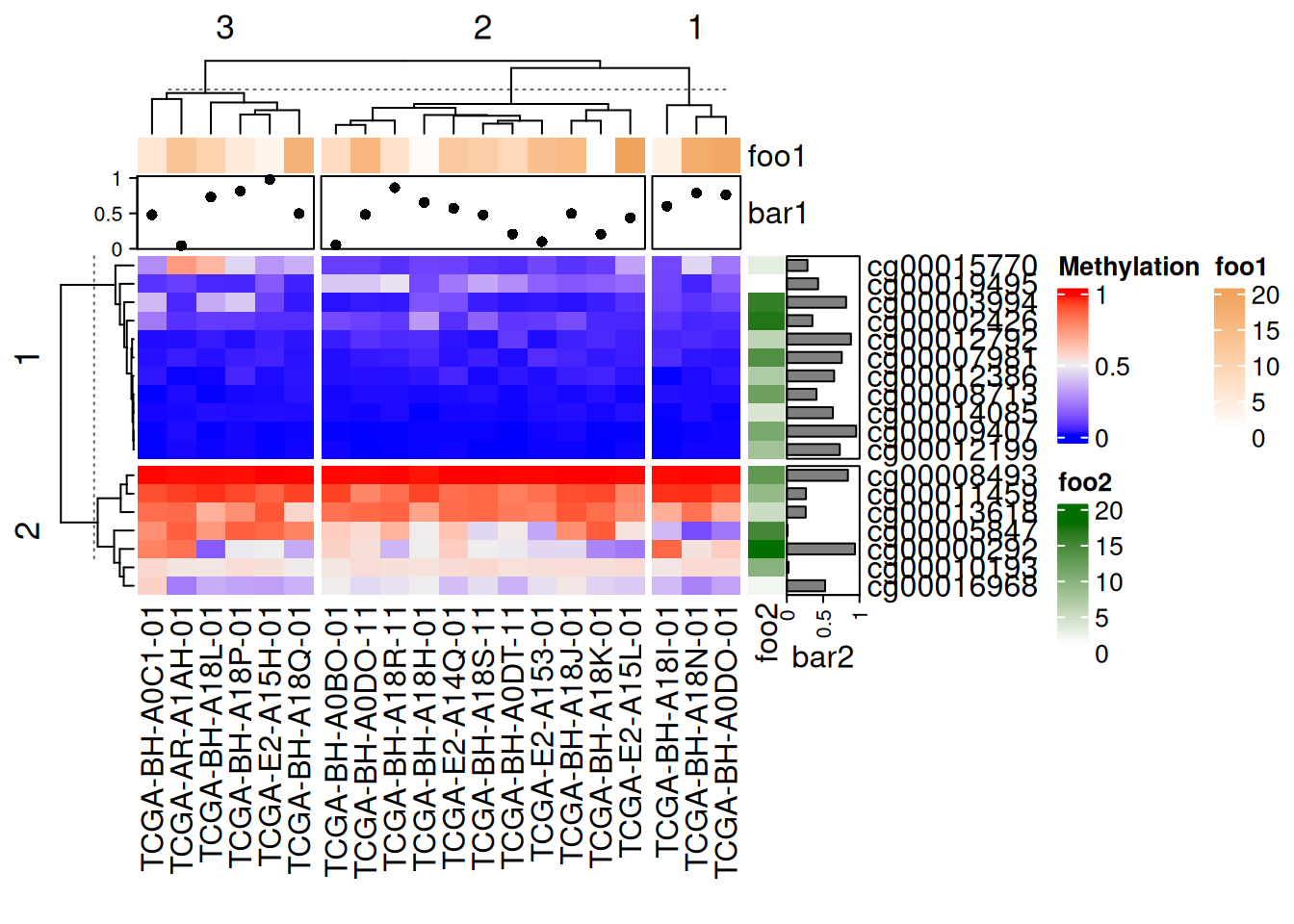
上图中添加的点图、热图、条图注释均随主热图分割
5. 标签添加
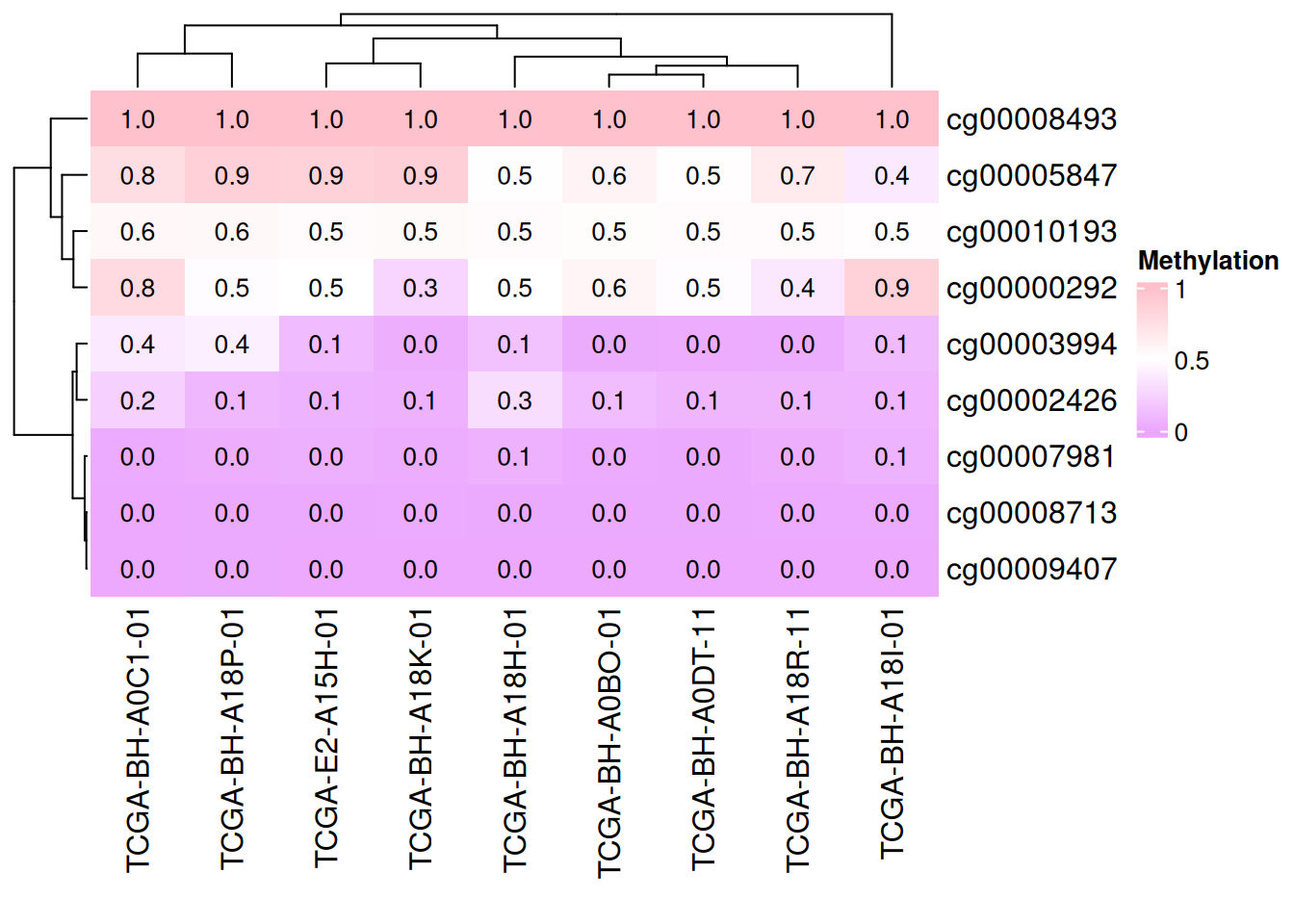
添加数据
cell_fun 接受一个函数,该函数会被应用到热图的每个单元格上。函数的参数如下:
j: 列索引(列号)。i: 行索引(行号)。x,y: 单元格中心点的坐标(基于绘图设备的坐标系)。width,height: 单元格的宽度和高度。fill: 单元格对应的数值(即矩阵中的值)。
# 添加数据
small_mat = data_TCGA[1:9, 1:9]
col_fun = colorRamp2(c(0, 0.5, 1), c("#EBA7FE", "white", "pink"))
Heatmap(small_mat, name = "Methylation", col = col_fun,
cell_fun = function(j, i, x, y, width, height, fill) {
grid.text(sprintf("%.1f", small_mat[i, j]), x, y, gp = gpar(fontsize = 10))
})
# 添加数据
small_mat = data_TCGA[1:9, 1:9]
Heatmap(small_mat, name = "Methylation", col = col_fun,
cell_fun = function(j, i, x, y, width, height, fill) {
if(small_mat[i, j] > 0.5) #设定显示范围
grid.text(sprintf("%.1f", small_mat[i, j]), x, y, gp = gpar(fontsize = 10)) #添加文本/数值
})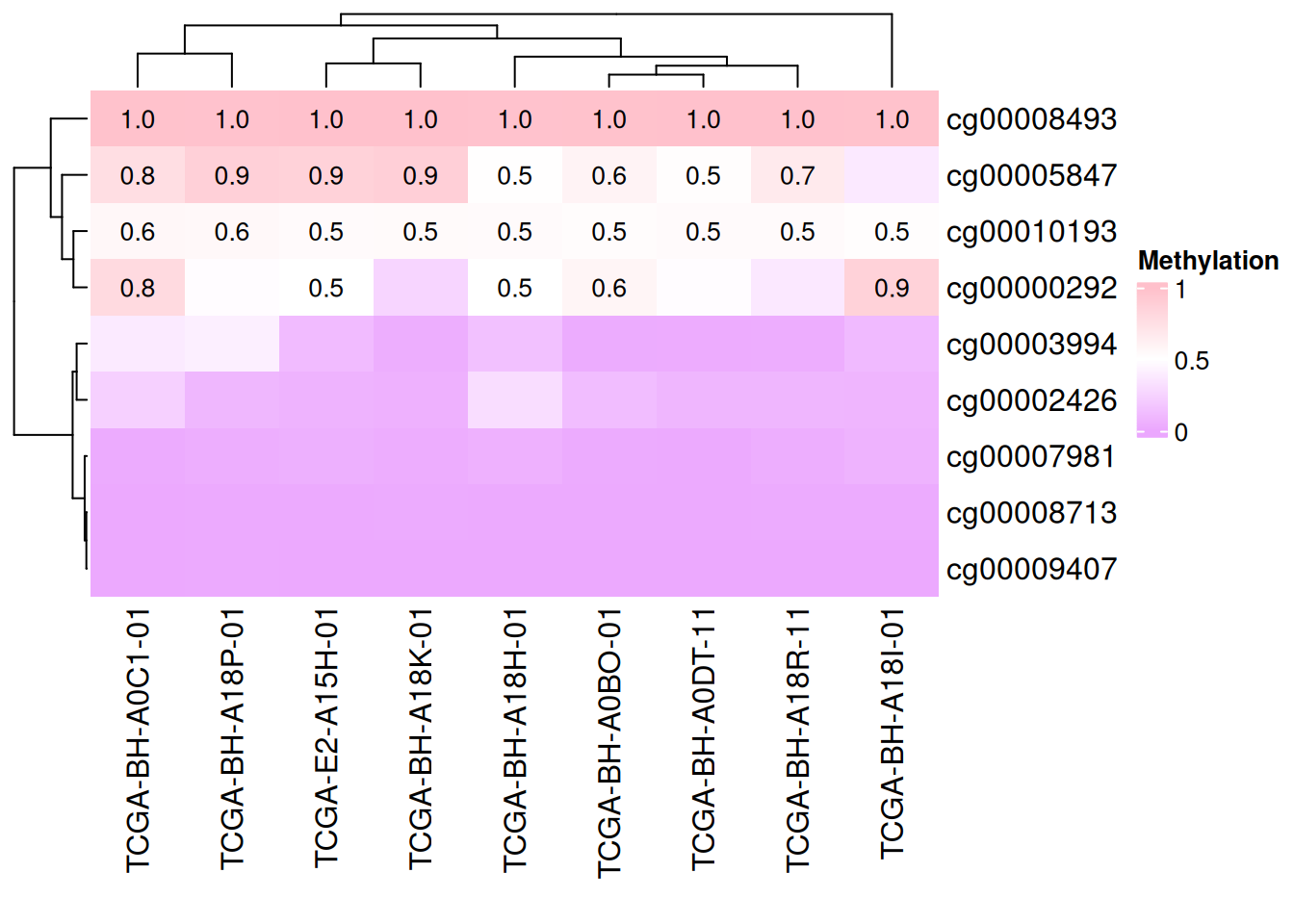
上图仅显示甲基化水平大于0.5的数据
添加图形
# 添加图形
small_mat = data_TCGA[1:9, 1:9]
Heatmap(small_mat, name = "Methylation", col = col_fun,
row_km = 2, column_km = 2,
layer_fun = function(j, i, x, y, w, h, fill) {
ind_mat = restore_matrix(j, i, x, y)
ind = unique(c(ind_mat[1, 4], ind_mat[3, 2])) #定位,每个分割对应位置添加图形
grid.points(x[ind], y[ind], pch = 16, size = unit(4, "mm")) #添加点
}
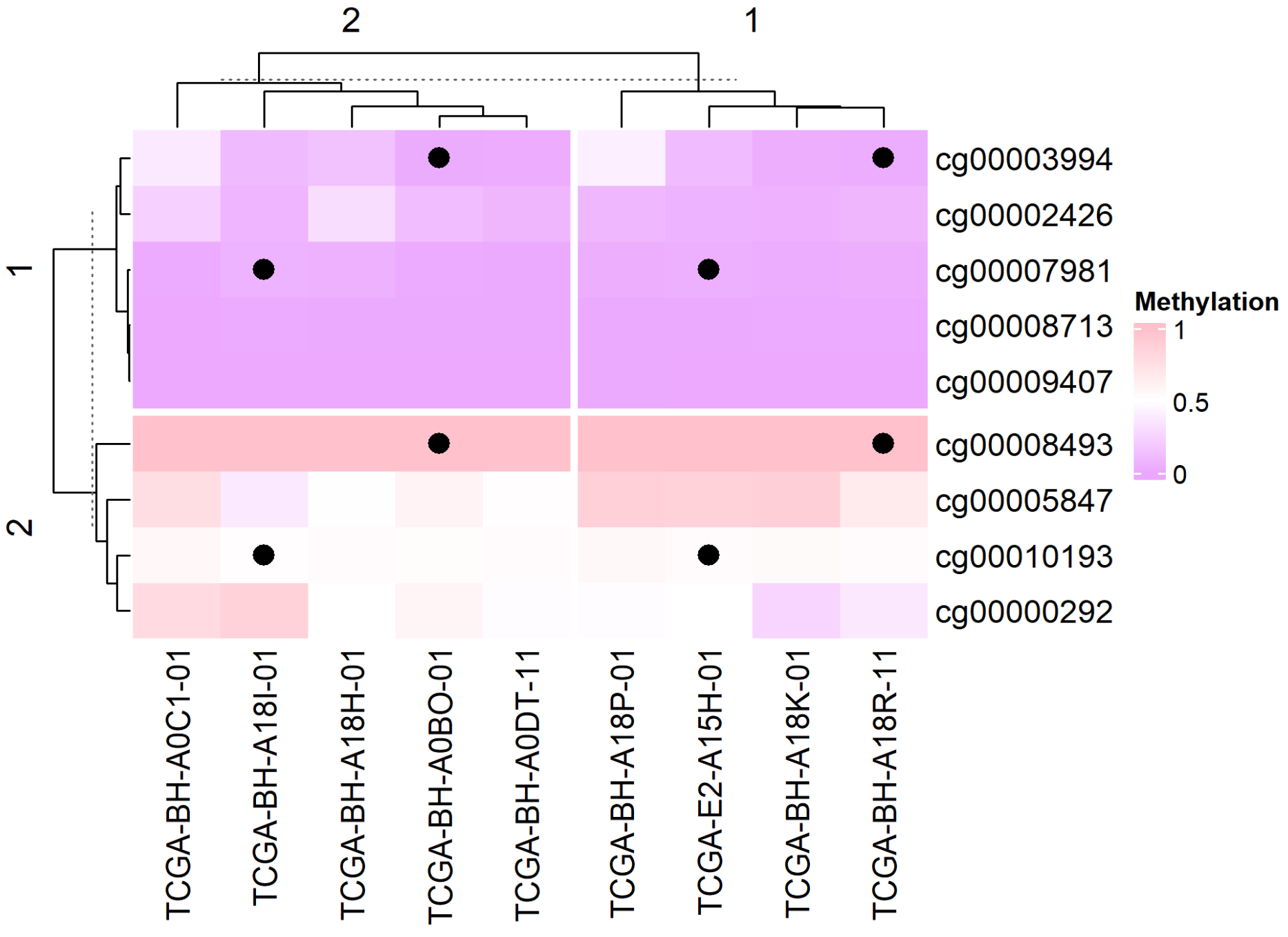
)
应用场景
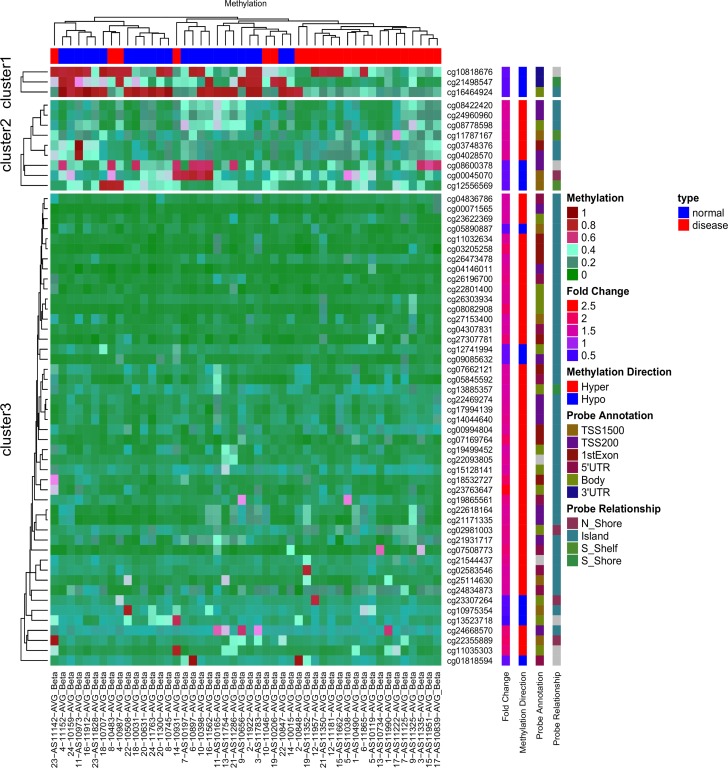
上图是基于 AVR 数据[1]中 59 个差异甲基化 ATG 位点的无监督层次聚类热图。红色表示 DNA 甲基化程度高,绿色表示甲基化程度低。
参考文献
[1] Radhakrishna U, Albayrak S, Alpay-Savasan Z, Zeb A, Turkoglu O, Sobolewski P, Bahado-Singh RO. Genome-Wide DNA Methylation Analysis and Epigenetic Variations Associated with Congenital Aortic Valve Stenosis (AVS). PLoS One. 2016 May 6;11(5):e0154010. doi: 10.1371/journal.pone.0154010. PMID: 27152866; PMCID: PMC4859473.
[2] Gu, Z. (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics.Gu, Z. (2022) Complex Heatmap Visualization. iMeta.
[3] Tal Galili (2015). dendextend: an R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics. DOI:10.1093/bioinformatics/btv428
[4] Wickham H, Vaughan D, Girlich M (2024). tidyr: Tidy Messy Data. R package version 1.3.1, CRAN: Package tidyr.
[5] Gu, Z. (2014) circlize implements and enhances circular visualization in R. Bioinformatics.
[6] Auguie B (2017). gridExtra: Miscellaneous Functions for “Grid” Graphics. R package version 2.3, CRAN: Package gridExtra.
[7] Kolde R (2019). pheatmap: Pretty Heatmaps. R package version 1.0.12, CRAN: Package pheatmap.
