# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("ComplexHeatmap", quietly = TRUE)) {
BiocManager::install("ComplexHeatmap")
}
# 加载包
library(data.table)
library(jsonlite)
library(ComplexHeatmap)大样本相关性热图
注记
Hiplot 网站
本页面为 Hiplot Corrplot Big Data 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
相关性热图是一种分析多个变量,两两之间相关性的图形。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;ComplexHeatmap
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-28
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
ComplexHeatmap * 2.26.0 2025-10-29 [1] Bioconduc~
data.table * 1.18.0 2025-12-24 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
载入数据为基因名称及每个样本的表达量。
# 加载数据
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/big-corrplot/data.json")$exampleData$textarea[[1]])
data <- as.data.frame(data)
# 整理数据格式
data <- data[!is.na(data[, 1]), ]
idx <- duplicated(data[, 1])
data[idx, 1] <- paste0(data[idx, 1], "--dup-", cumsum(idx)[idx])
rownames(data) <- data[, 1]
data <- data[, -1]
str2num_df <- function(x) {
x[] <- lapply(x, function(l) as.numeric(l))
x
}
tmp <- t(str2num_df(data))
corr <- round(cor(tmp, use = "na.or.complete", method = "pearson"), 3)
# 查看数据
head(corr) RGL4 MPP7 UGCG CYSTM1 ANXA2 ENDOD1 ARHGAP24 CST7 HIST1H2BM
RGL4 1.000 0.914 0.929 0.936 -0.592 -0.908 0.888 0.949 -0.603
MPP7 0.914 1.000 0.852 0.907 -0.543 -0.862 0.762 0.899 -0.656
UGCG 0.929 0.852 1.000 0.956 -0.440 -0.791 0.854 0.840 -0.694
CYSTM1 0.936 0.907 0.956 1.000 -0.358 -0.762 0.812 0.852 -0.632
ANXA2 -0.592 -0.543 -0.440 -0.358 1.000 0.826 -0.660 -0.723 0.541
ENDOD1 -0.908 -0.862 -0.791 -0.762 0.826 1.000 -0.907 -0.961 0.709
EREG EMP1 NFAM1 SLC40A1 CD52 HIST1H2BH PFKFB3 SNORD116-20 STX11
RGL4 -0.021 -0.495 0.859 0.506 -0.704 -0.680 0.889 0.188 0.953
MPP7 -0.196 -0.447 0.898 0.648 -0.734 -0.770 0.842 0.048 0.915
UGCG 0.153 -0.358 0.858 0.361 -0.671 -0.711 0.943 0.202 0.951
CYSTM1 0.074 -0.272 0.866 0.339 -0.612 -0.683 0.933 0.225 0.985
ANXA2 0.222 0.902 -0.662 -0.668 0.775 0.626 -0.463 0.375 -0.374
ENDOD1 0.191 0.713 -0.872 -0.611 0.854 0.791 -0.814 0.141 -0.787
SYNE2 TCN1
RGL4 0.780 0.889
MPP7 0.795 0.888
UGCG 0.922 0.927
CYSTM1 0.908 0.973
ANXA2 -0.327 -0.249
ENDOD1 -0.657 -0.708可视化
# 大样本相关性热图
p <- ComplexHeatmap::Heatmap(
corr, col = colorRampPalette(c("#4477AA","#FFFFFF","#BB4444"))(50),
clustering_distance_rows = "euclidean",
clustering_method_rows = "ward.D2",
clustering_distance_columns = "euclidean",
clustering_method_columns = "ward.D2",
show_column_dend = FALSE, show_row_dend = FALSE,
column_names_gp = gpar(fontsize = 8),
row_names_gp = gpar(fontsize = 8)
)
p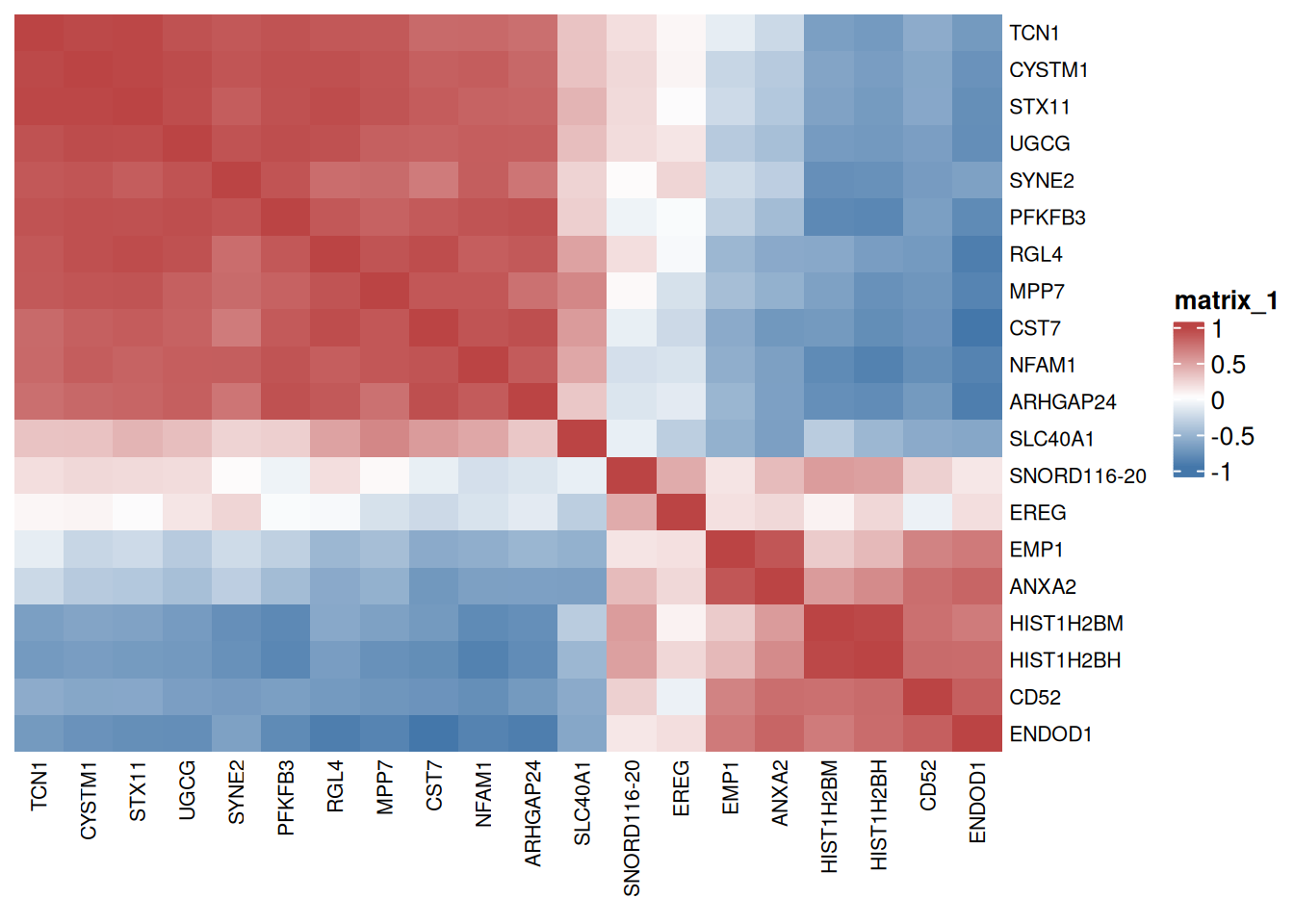
红色系表示两个基因之间呈正相关,蓝色系表示两个基因之间呈负相关,每一格中的数字表示相关系数。
