# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("ggrepel", quietly = TRUE)) {
install.packages("ggrepel")
}
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("RColorBrewer", quietly = TRUE)) {
install.packages("RColorBrewer")
}
# 加载包
library(data.table)
library(jsonlite)
library(ggrepel)
library(ggplot2)
library(RColorBrewer)基因差异表达排序图
注记
Hiplot 网站
本页面为 Hiplot Gene Ranking Dotplot 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
基因表达排序可视化。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;ggrepel;ggplot2;RColorBrewer
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-28
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.0 2025-12-24 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
ggrepel * 0.9.6 2024-09-07 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
RColorBrewer * 1.1-3 2022-04-03 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
# 加载数据
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/gene-rank/data.json")$exampleData$textarea[[1]])
data <- as.data.frame(data)
# 整理数据格式
## 按 log2FoldChange 和 pvalue 排序
data <- data[order(-data$log2FC, data$pvalue), ]
## 添加 rank 列
data$rank <- 1:nrow(data)
## 获取 top n 上调和下调基因进行标记
top_n <- 5
top_n_up <- rownames(head(data, top_n))
top_n_down <- rownames(tail(data, top_n))
genes_to_label <- c(top_n_up, top_n_down)
data2 <- data[genes_to_label, ]
# 查看数据
head(data) gene log2FC pvalue rank
514 LOC91370 3.889820 8.478310e-03 1
293 LRRC25 3.492413 5.000000e-05 2
470 BEGAIN 3.312996 3.347634e-03 3
159 RDM1P5 3.125070 1.538852e-02 4
194 CLDN1 3.096516 2.622163e-02 5
626 KCNS1 2.949729 2.770000e-09 6可视化
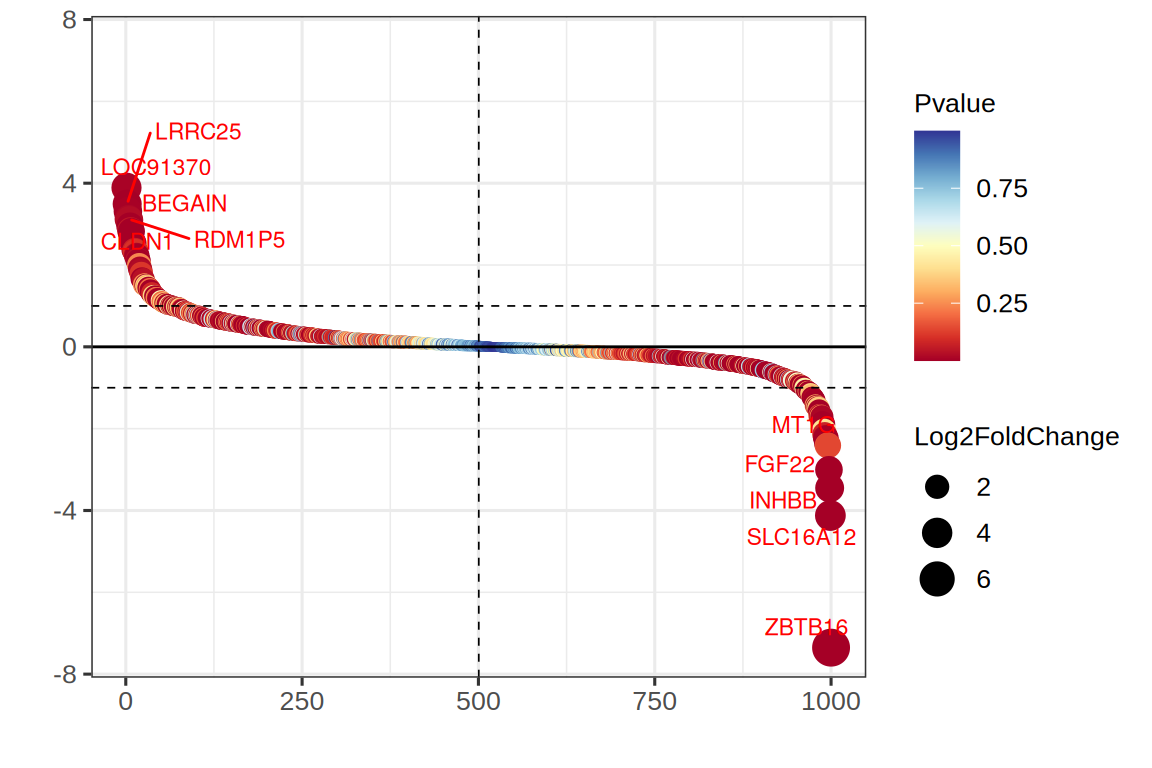
# 基因差异表达排序图
p <-
ggplot(data, aes(rank, log2FC, color = pvalue, size = abs(log2FC))) +
geom_point() +
scale_color_gradientn(colours = colorRampPalette(brewer.pal(11,'RdYlBu'))(100)) +
geom_hline(yintercept = c(-1, 1), linetype = 2, size = 0.3) +
geom_hline(yintercept = 0, linetype = 1, size = 0.5) +
geom_vline(xintercept = median(data$rank), linetype = 2, size = 0.3) +
geom_text_repel(data = data2, aes(rank, log2FC, label = gene),
size = 3, color = "red") +
xlab("") + ylab("") +
ylim(c(-max(abs(data$log2FC)), max(abs(data$log2FC)))) +
labs(color = "Pvalue", size = "Log2FoldChange") +
theme_bw(base_size = 12) +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p