# 安装包
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("ggseqlogo", quietly = TRUE)) {
install.packages("ggseqlogo")
}
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
# 加载包
library(data.table)
library(jsonlite)
library(ggseqlogo)
library(ggplot2)序列 Logo
注记
Hiplot 网站
本页面为 Hiplot Seqlogo 插件的源码版本教程,您也可以使用 Hiplot 网站实现无代码绘图,更多信息请查看以下链接:
序列 Logo 是一种用来描述结合位点序列模式的图形。
环境配置
系统: Cross-platform (Linux/MacOS/Windows)
编程语言: R
依赖包:
data.table;jsonlite;ggseqlogo;ggplot2
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-27
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.0 2025-12-24 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
ggseqlogo * 0.2.2 2025-12-22 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
载入数据为多个转录因子在多个基因上的结合位点的序列。
# 加载数据
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/ggseqlogo/data.json")$exampleData$textarea[[1]])
data <- as.data.frame(data)
# 整理数据格式
data <- data[, !sapply(data, function(x) {all(is.na(x))})]
data <- as.list(data)
data <- lapply(data, function(x) {return(x[!is.na(x)])})
# 查看数据
str(data[1:5])List of 5
$ MA0001.1: chr [1:97] "CCATATATAG" "CCATATATAG" "CCATAAATAG" "CCATAAATAG" ...
$ MA0002.1: chr [1:26] "AATTGTGGTTA" "ATCTGTGGTTA" "AATTGTGGTAA" "TTCTGCGGTTA" ...
$ MA0004.1: chr [1:20] "CACGTG" "CACGTG" "CACGTG" "CACGTG" ...
$ MA0005.1: chr [1:90] "CCTAATTGGGC" "CCTAATTTGGC" "CCTAATCGGGC" "CCTAATCGGGC" ...
$ MA0006.1: chr [1:24] "CGCGTG" "CGCGTG" "CGCGTG" "CGCGTG" ...可视化
# 序列 Logo
p <- ggseqlogo(
data,
ncol = 4,
col_scheme = "nucleotide",
seq_type = "dna",
method = "bits") +
theme(plot.title = element_text(hjust = 0.5))
p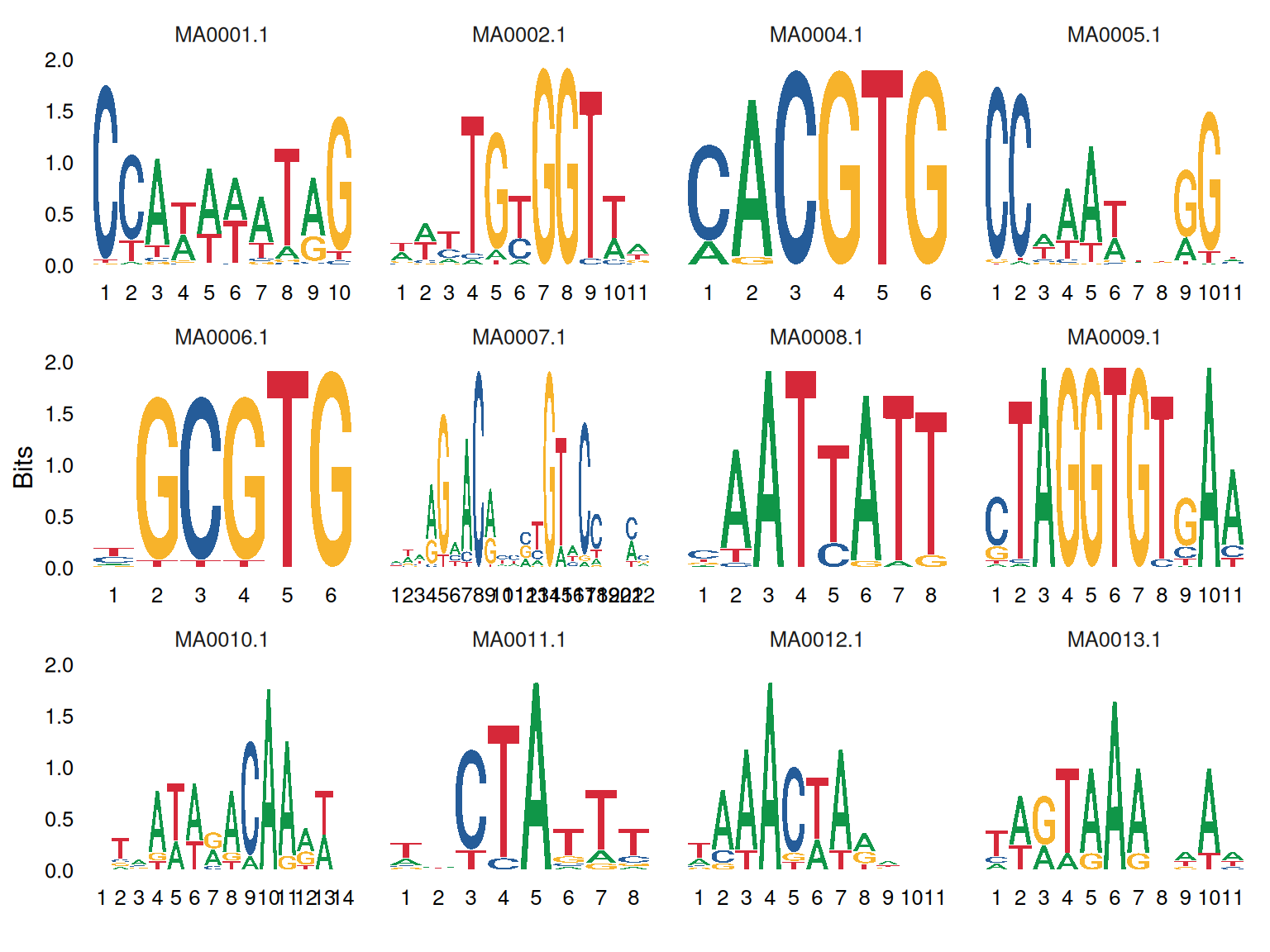
采用 bits 计算的方式将一个结合位点序列展示在图表一列,可以清晰观测到不同序列占比较大的碱基。
