# 安装包
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("dplyr", quietly = TRUE)) {
install.packages("dplyr")
}
if (!requireNamespace("scales", quietly = TRUE)) {
install.packages("scales")
}
if (!requireNamespace("ggforce", quietly = TRUE)) {
install.packages("ggforce")
}
# 加载包
library(ggplot2)
library(dplyr)
library(scales)
library(ggforce)径向柱状图
示例
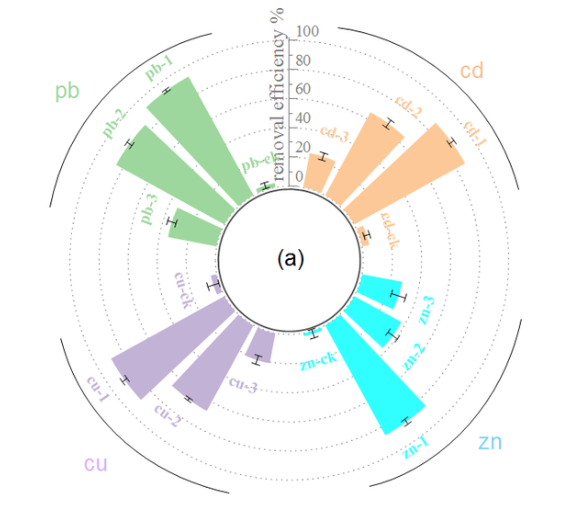
通过环形布局展示四足动物各分类群受威胁物种比例,蓝色基准线表示最佳估计比例,灰色网格线辅助百分比读数。
环境配置
系统要求: 跨平台(Linux/MacOS/Windows)
编程语言:R
依赖包:
ggplot2,dplyr,scales,ggforce
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-04
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
dplyr * 1.1.4 2023-11-17 [1] RSPM
ggforce * 0.5.0 2025-06-18 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
scales * 1.4.0 2025-04-24 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
- 需包含R内置的数据集(如iris、penguins)和生物医学相关数据集(如组学数据、生存信息、临床指标等)。
- 生物医学相关数据集需上传至Bizard腾讯云以便获取插入教程的链接,来自于公共数据集的数据最佳,若由个人/组织提供需确保数据能够被公开。数据集大小需小于1MB。
# 数据读入和处理代码可以自由选择是否展示------
# 生成模拟临床数据
set.seed(123)
n <- 12 # 样本量
df <- data.frame(
id = 1:n,
patient = paste0("P-", sprintf("%02d", 1:n)),
value = c(rnorm(6, 80, 15), rnorm(6, 120, 20)), # 对照组与治疗组
group = rep(c("Control", "Treatment"), each = 6)
) %>%
mutate(
angle = 90 - 360 * (id - 0.5)/n,
hjust = ifelse(angle < -90, 1, 0),
angle = ifelse(angle < -90, angle + 180, angle)
)
# 添加内置数据集
data("iris")可视化
1. 简单环形布局
# 基础柱状图
p1 <- ggplot(df, aes(x = factor(id), y = value)) +
geom_col(aes(fill = group), width = 0.8, alpha = 0.8) +
coord_radial(inner.radius = 0.3) +
scale_fill_manual(values = c("#1E88E5", "#D81B60")) +
theme_void() +
labs(title = "治疗组与对照组指标对比")
p1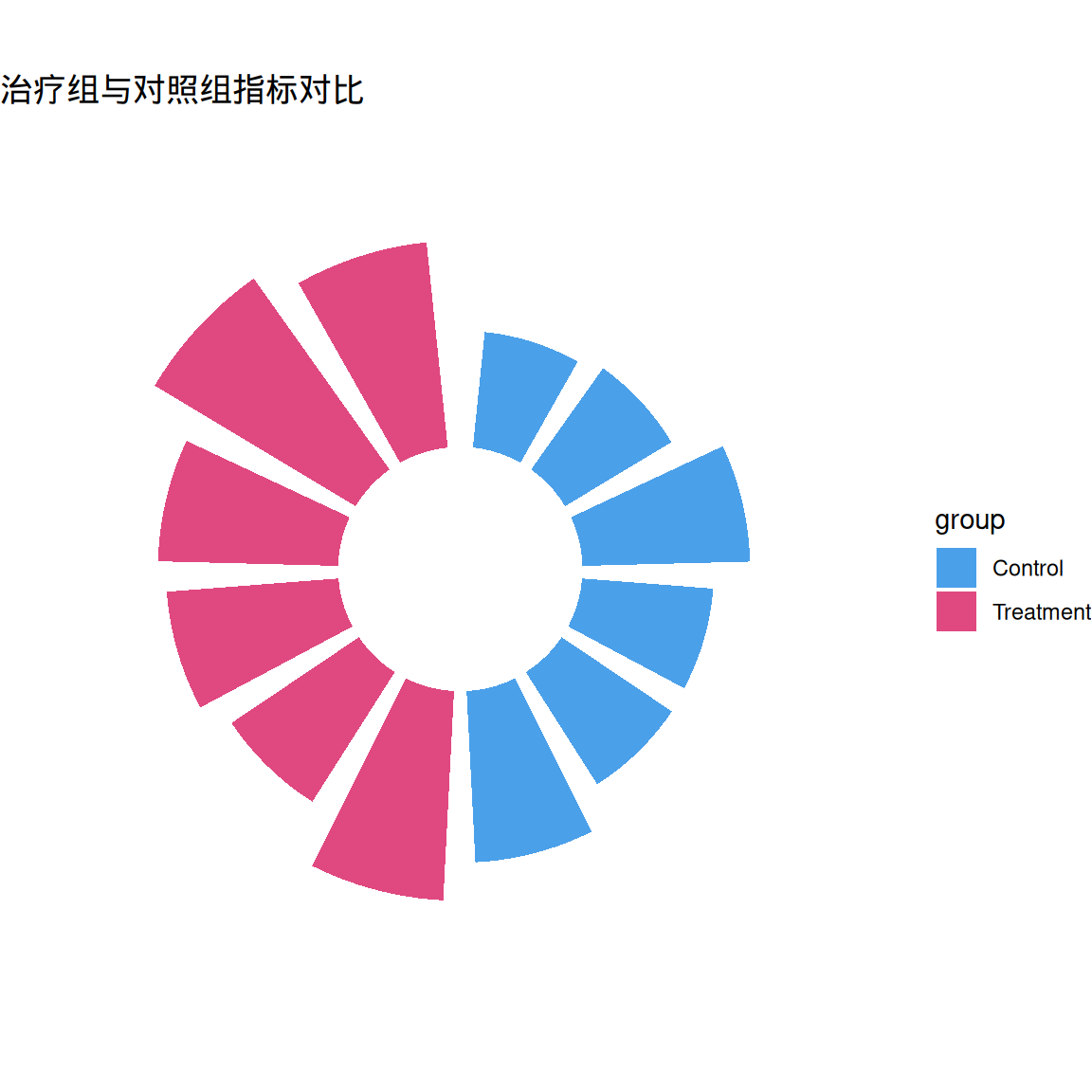
补充基础代码可以扩展的重要参数,并提供对应的绘图代码,例如:
提示
关键参数:
coord_radial(inner.radius=0.3): 控制中心空白区域比例width=0.8: 柱宽建议值范围0.5-1.0alpha=0.8: 设置颜色透明度
p2 <- ggplot(df, aes(x = factor(id))) +
geom_col(aes(y = value, fill = group), width = 0.85) +
geom_text(aes(y = value + 15, label = patient, angle = angle),
size = 3, hjust = 0.3) +
coord_radial(start = -0.05 * pi) +
scale_fill_manual(values = c("#004D40", "#FFA000")) +
theme_void() +
labs(fill = "实验分组")
p2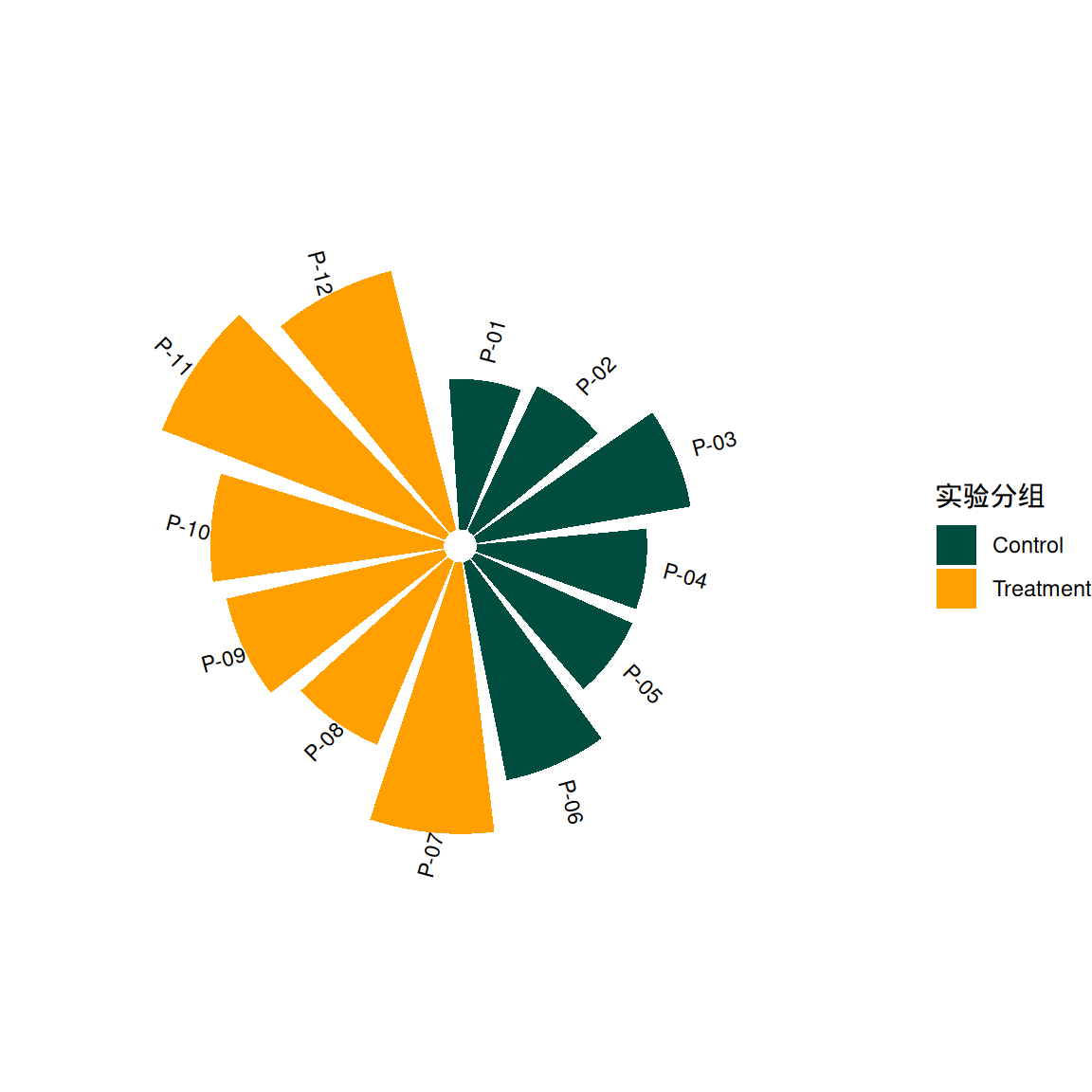
2. 更多进阶图表
p1 <- ggplot(df) +
geom_col(aes(x = id, y = value, fill = group), width = 0.85) +
geom_hline(yintercept = c(50, 100, 150),
color = "grey80", linetype = "dashed") +
geom_segment(aes(x = 0.5, xend = n+0.5, y = 100, yend = 100),
color = "#283593", linewidth = 0.8) +
geom_text(aes(x = id, y = value + 20, label = patient, angle = angle),
size = 3, hjust = 0.3) +
coord_radial(inner.radius = 0.25) +
scale_fill_brewer(palette = "Set2") +
theme_void() +
labs(caption = "虚线表示临床指标参考区间") +
theme(legend.position = "bottom")
p1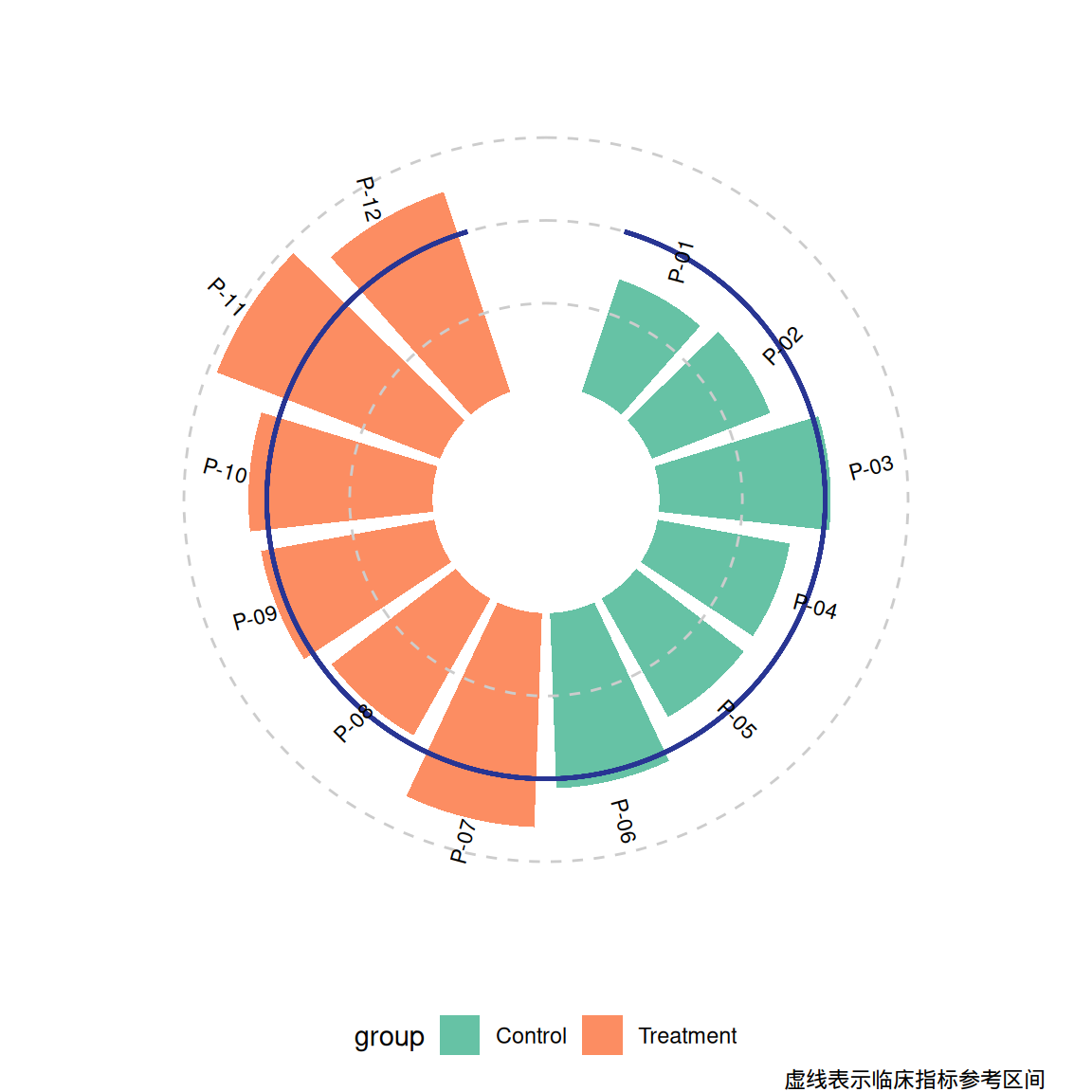
# 生成主绘图对象
p1 <- ggplot(df) +
geom_col(aes(x = id, y = value, fill = group),
width = 0.85, alpha = 0.8) +
# ========== 核心坐标轴系统 ==========
# 主Y轴线(沿0度方向)
geom_segment(aes(x = 0.5, xend = 0.5, y = 0, yend = max(df$value) * 1.1),
color = "grey40", linewidth = 0.6,
arrow = arrow(length = unit(0.2, "cm"))) +
# Y轴刻度线(放射状)
do.call(c, lapply(c(50, 100, 150), function(yval) {
geom_segment(aes(x = 0.5 - 0.05, xend = 0.5 + 0.05,
y = yval, yend = yval),
color = "grey50", linewidth = 0.4)
})) + # 合并列表中的元素
# Y轴刻度标签(沿轴线方向)
do.call(c, lapply(c(50, 100, 150), function(yval) {
geom_text(aes(x = 0.5 + 0.1, y = yval,
label = yval, angle = -90),
color = "grey30", size = 3.5, hjust = 0)
})) + # 合并列表中的元素
# 其他横线和数据标签
geom_hline(yintercept = c(50, 100, 150),
color = "grey80", linetype = "dashed") +
geom_segment(aes(x = 0.5, xend = 0.5, y = 100, yend = 100),
color = "#283593", linewidth = 0.8) + # 修改 end 坐标,以适应极坐标
geom_text(aes(x = id, y = value + 20, label = patient, angle = angle),
size = 3, hjust = 0.3) +
# ========== 坐标系设置 ==========
coord_radial(inner.radius = 0.35,
start = -0.1 * pi) +
# ========== 视觉映射 ==========
scale_fill_viridis_d(option = "C", begin = 0.2) +
theme_void() +
labs(caption = "虚线表示临床指标参考区间") +
theme(plot.margin = margin(2, 2, 2, 2, "cm"),
legend.position = "bottom")
p1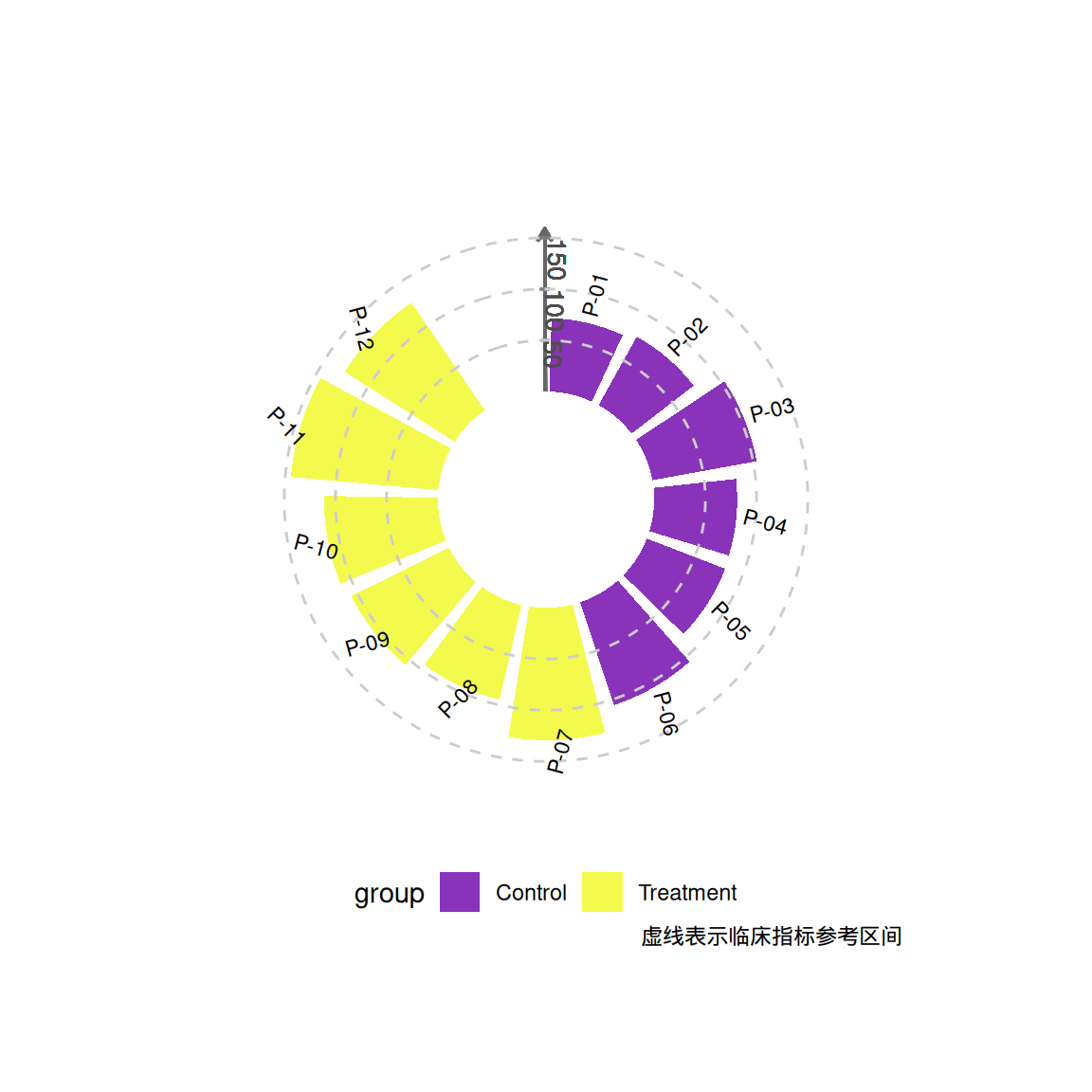
应用场景
展示可视化图表在生物医学文献中的实际应用,如果基础图表/进阶图表被广泛应用在各类生物医学文献,则可以选择分别展示。
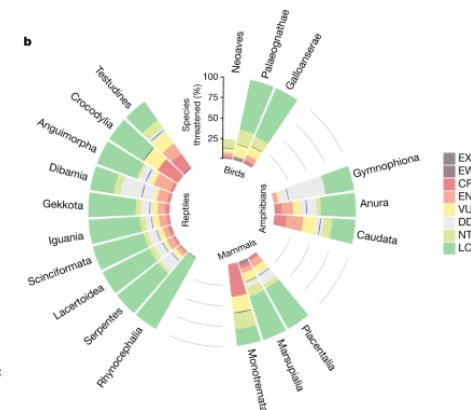
该径向柱状图通过环形布局直观展示四足动物保护现状
参考文献
[1] Cox N, Young BE, & Xie Y. A global reptile assessment highlights shared conservation needs of tetrapods. Nature. 2022 May;605(7909):285-290. https://doi.org/10.1038/s41586-022-04664-7
[2] Costa, A. M., Machado, J. T., & Quelhas, M. D. (2011). Histogram-based DNA analysis for the visualization of chromosome, genome, and species information. Bioinformatics, 27(9), 1207–1214. https://doi.org/10.1093/bioinformatics/btr131
