# 安装包
if (!requireNamespace("treemap", quietly = TRUE)) {
install.packages("treemap")
}
if (!requireNamespace("tidyverse", quietly = TRUE)) {
install.packages("tidyverse")
}
if (!requireNamespace("stats", quietly = TRUE)) {
install.packages("stats")
}
if (!requireNamespace("DOSE", quietly = TRUE)) {
install.packages("DOSE")
}
# 加载包
library(treemap)
library(tidyverse)
library(stats)
library(DOSE)
library(palmerpenguins)矩形树图
矩形树图(Treemap)也叫矩形式树状结构图,它采用多组面积不等的矩形嵌套而成。所有矩形的面积之和代表了总体数据。各个小矩形的面积表示每个子项的占比,矩形面积越大,表示子数据在整体中的占比越大。
示例
矩形树图适合展现具有层级关系的数据,能够直观体现同级之间的比较(矩形树图使用不同颜色和大小的长方形来显示数据的层次结构。
环境配置
系统要求: 跨平台(Linux/MacOS/Windows)
编程语言:R
依赖包:
treemap;tidyverse;stats;DOSE
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-02
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
DOSE * 4.4.0 2025-10-29 [1] Bioconduc~
dplyr * 1.1.4 2023-11-17 [1] RSPM
forcats * 1.0.1 2025-09-25 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
lubridate * 1.9.4 2024-12-08 [1] RSPM
palmerpenguins * 0.1.1 2022-08-15 [1] RSPM
purrr * 1.2.1 2026-01-09 [1] RSPM
readr * 2.1.6 2025-11-14 [1] RSPM
stringr * 1.6.0 2025-11-04 [1] RSPM
tibble * 3.3.1 2026-01-11 [1] RSPM
tidyr * 1.3.2 2025-12-19 [1] RSPM
tidyverse * 2.0.0 2023-02-22 [1] RSPM
treemap * 2.4-4 2023-05-25 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
data_USArrests <- rownames_to_column(USArrests[1:8,], "State")
data_swiss <- swiss
data_countsub <- aggregate(penguins, by=list(penguins$species, penguins$sex),length)
data_countsub <- data_countsub[ ,1:3]
colnames(data_countsub) <- c("species", "sex", "count")
data_BP <- readr::read_csv("https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/data_BP.csv")
data_BP <- data_BP[order(abs(data_BP$NES), decreasing = T),]
data_BP <- data_BP[1:13,]
data_KEGG_type <- readr::read_csv("https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/data_KEGG.csv")
data_KEGG_type$pvalue_log <- -log10(data_KEGG_type$pvalue)可视化
1. 单一变量分类
1.1 依据标签填色
以USArrests数据集为例
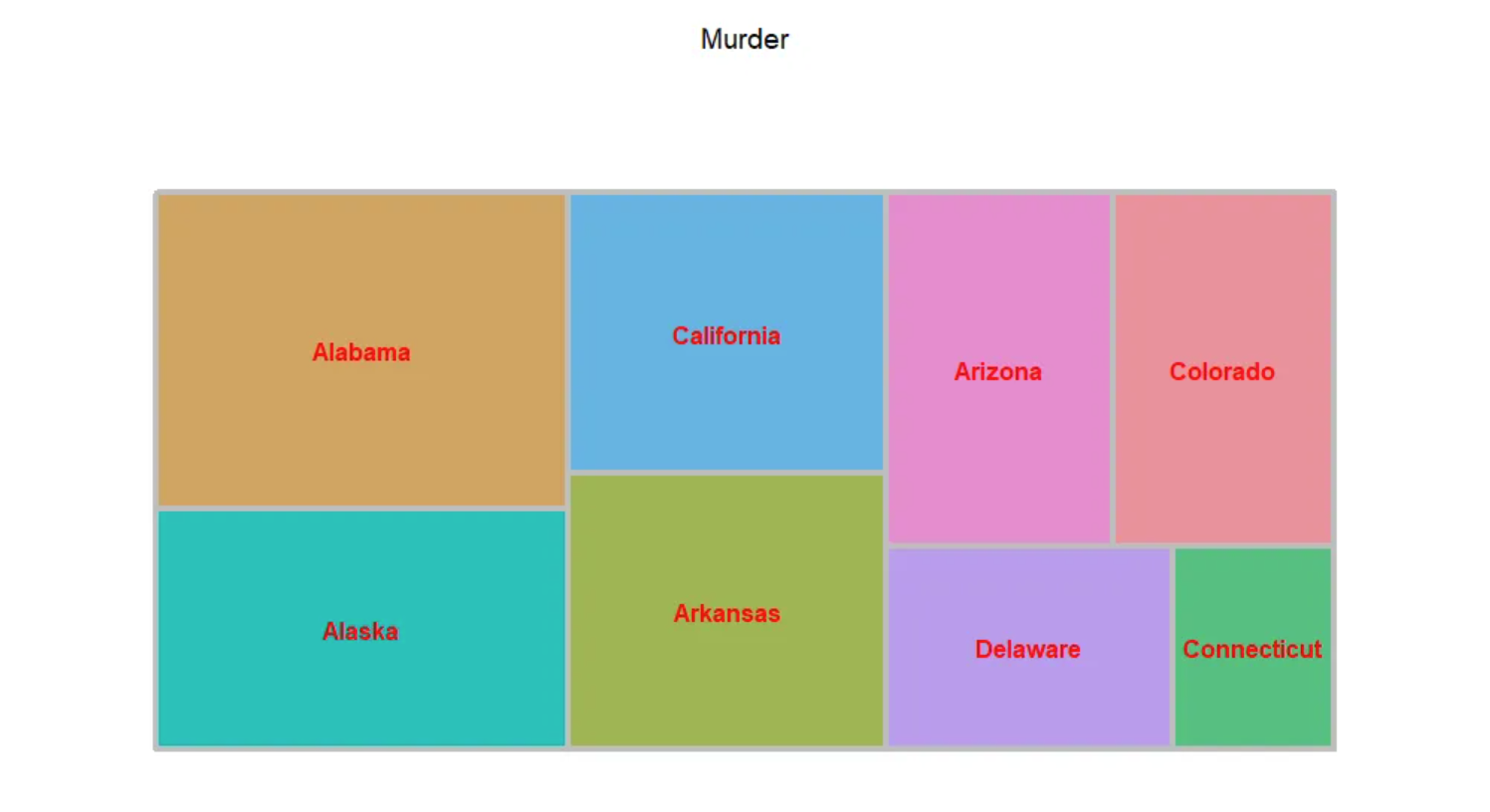
treemap(data_USArrests, # 数据
index = "State", # 分类变量
vSize = "Murder", # 分类变量对应数据值
vColor="State", # 颜色深浅的对应列,这里以数据大小作为对应
type = "index", # "颜色映射方式,"index"、"value"、"comp"、"dens"、"depth"、"categorical"、"color"、"manual"
title = 'Murder', # 标题
border.col = "grey", # 边框颜色
border.lwds = 4, # 边框线宽度
fontsize.labels = 12, # 标签大小
fontcolor.labels = 'red', # 标签颜色
align.labels = list(c("center", "center")), # 标签位置
fontface.labels = 2) # 标签字体:1,2,3,4 表示正常、粗体、斜体、粗斜体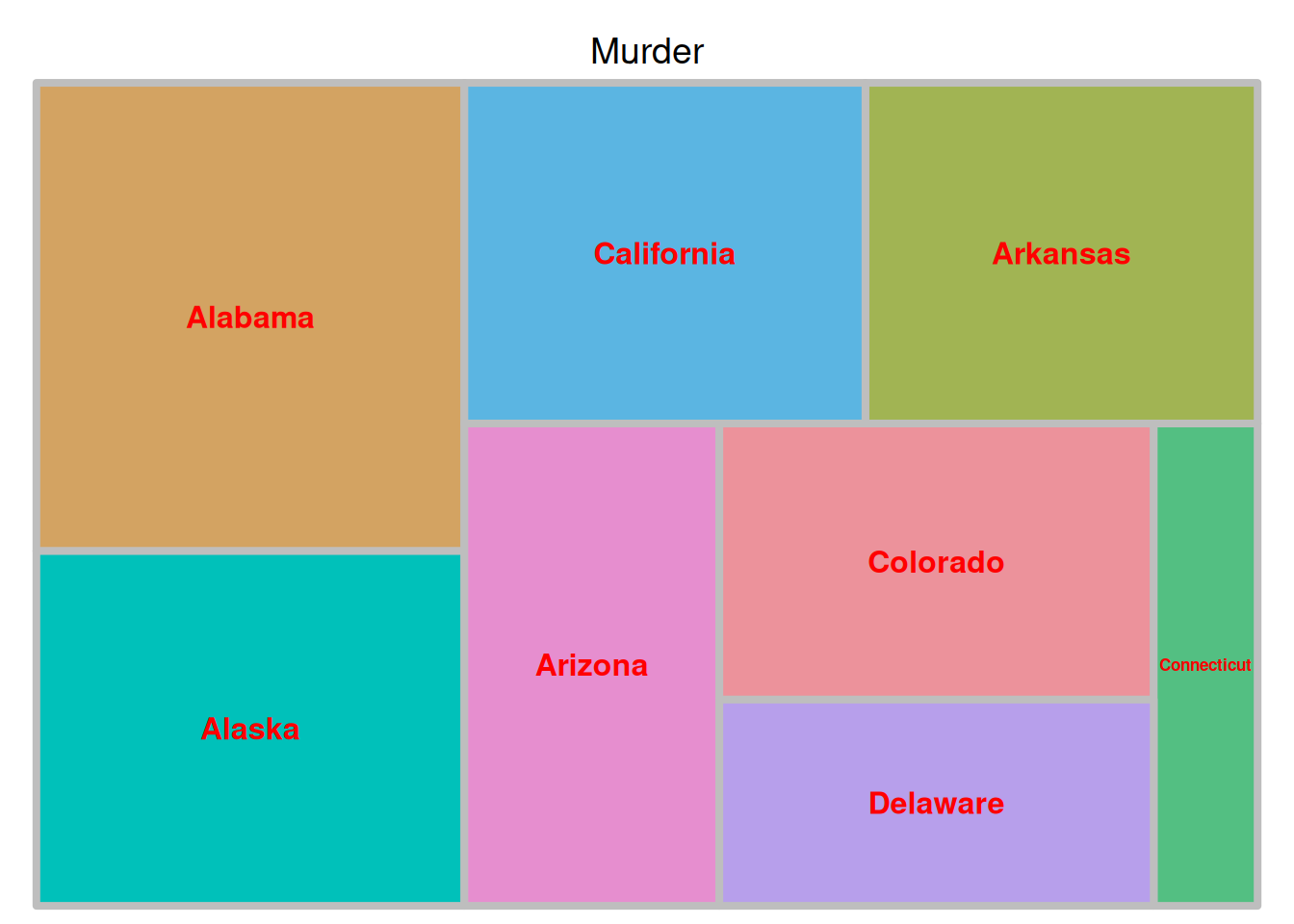
上图展示了美国不同州犯罪被捕人数比例的矩形树图,以州作为填色依据。
1.2 依据数据大小填色
以USArrests数据集为例
treemap(data_USArrests,
index = "State",
vSize = "Murder",
vColor="Murder",
type = "value",
title = 'Murder',
border.col = "grey",
border.lwds = 4,
fontsize.labels = 12,
fontcolor.labels = 'red',
align.labels = list(c("center", "center")),
fontface.labels = 2)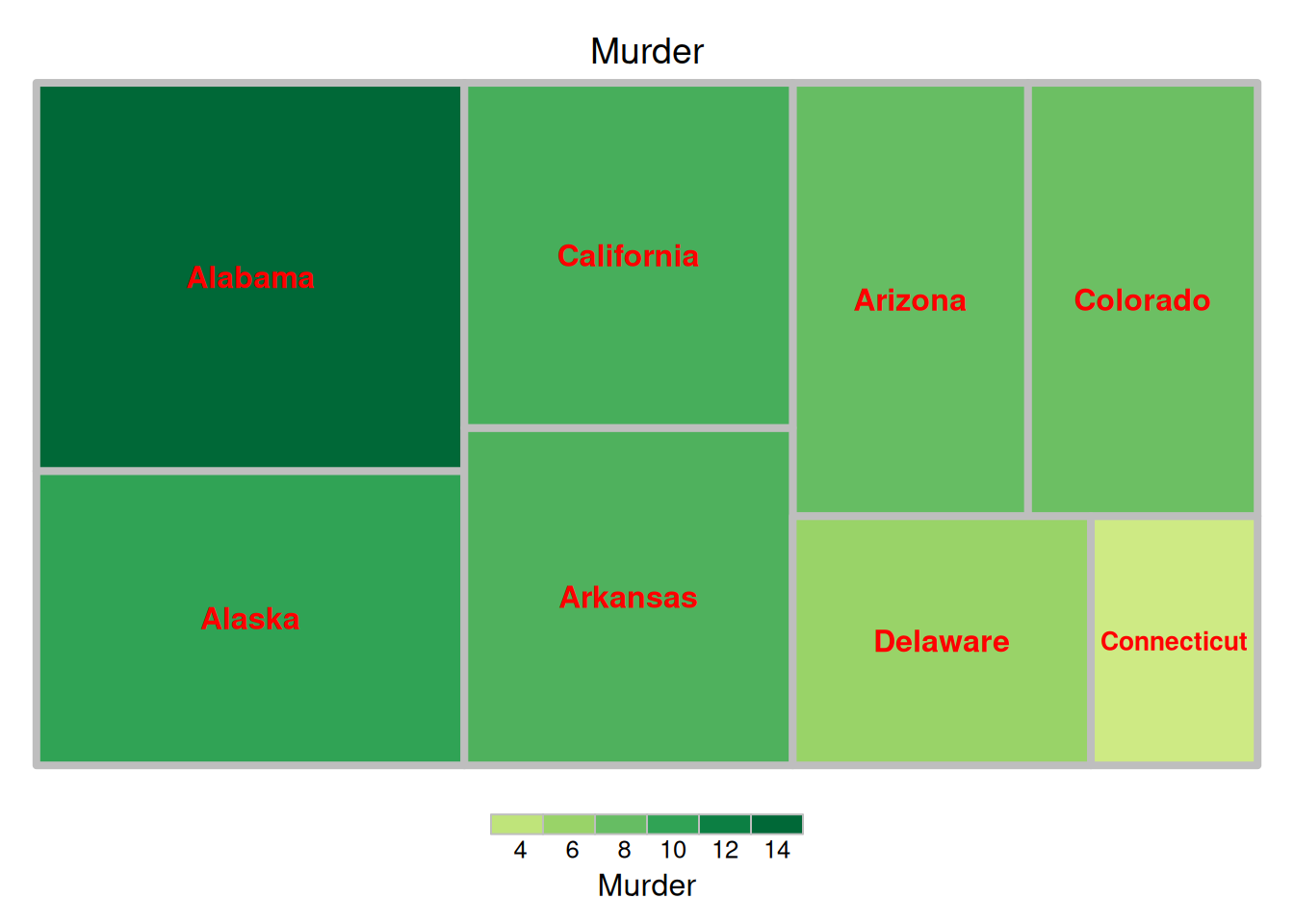
上图展示了美国不同州犯罪被捕人数比例的矩形树图,以各组数据大小作为填色依据。
以小鼠肺组织使用顺铂前后的差异基因对应的HALLMARK分析结果数据为例:
treemap(data_BP[1:13,],
index = "BP",
vSize = "pvalue_log",
vColor="NES",
type = "value",
title = 'Biological Process',
border.col = "grey",
border.lwds = 4,
fontsize.labels = 10,
fontcolor.labels = 'white',
align.labels = list(c("center", "center")),
fontface.labels = 2)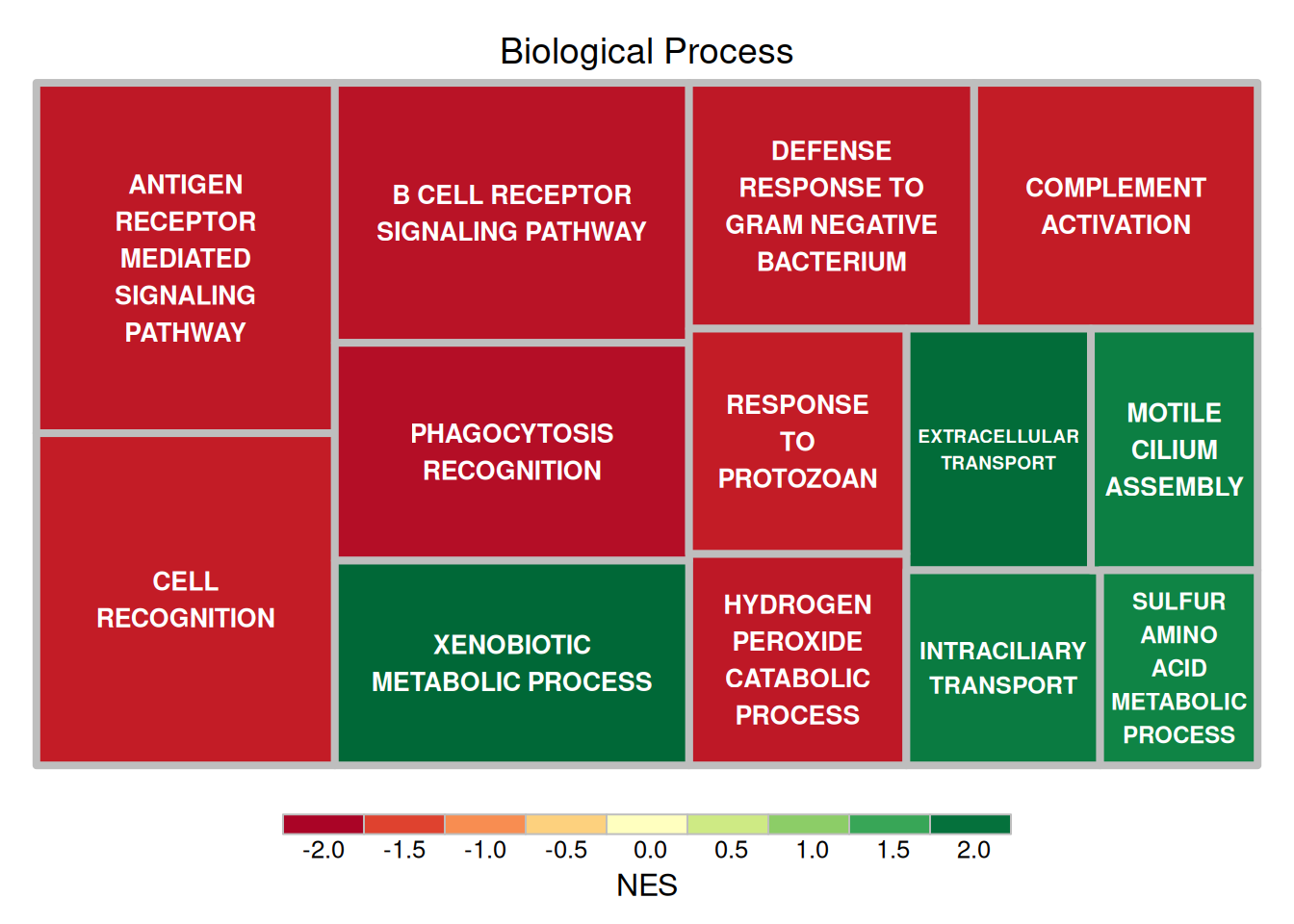
上图展示了小鼠肺组织使用顺铂前后的差异基因HALLMARK分析的结果,矩形大小表示log10(P)的负值,矩形越大表示结果越可靠。矩形颜色表示NES值。
2. 多变量分类
利用index = c("Group1”,"Group2", ...)实现多层级分组
2.1 依据数据标签填色
以penguins数据集为例
treemap(data_countsub,
index = c("sex","species"),
vSize = "count",
vColor="species",
type = "index",
title = 'Count',
border.col = c("black","white"),
border.lwds = c(4,1),
fontsize.labels = c(18,10),
bg.labels=c("transparent"),
fontcolor.labels = c('white',"orange"),
align.labels = list(c("left", "top"),
c("center", "center")),
fontface.labels = c(2,3))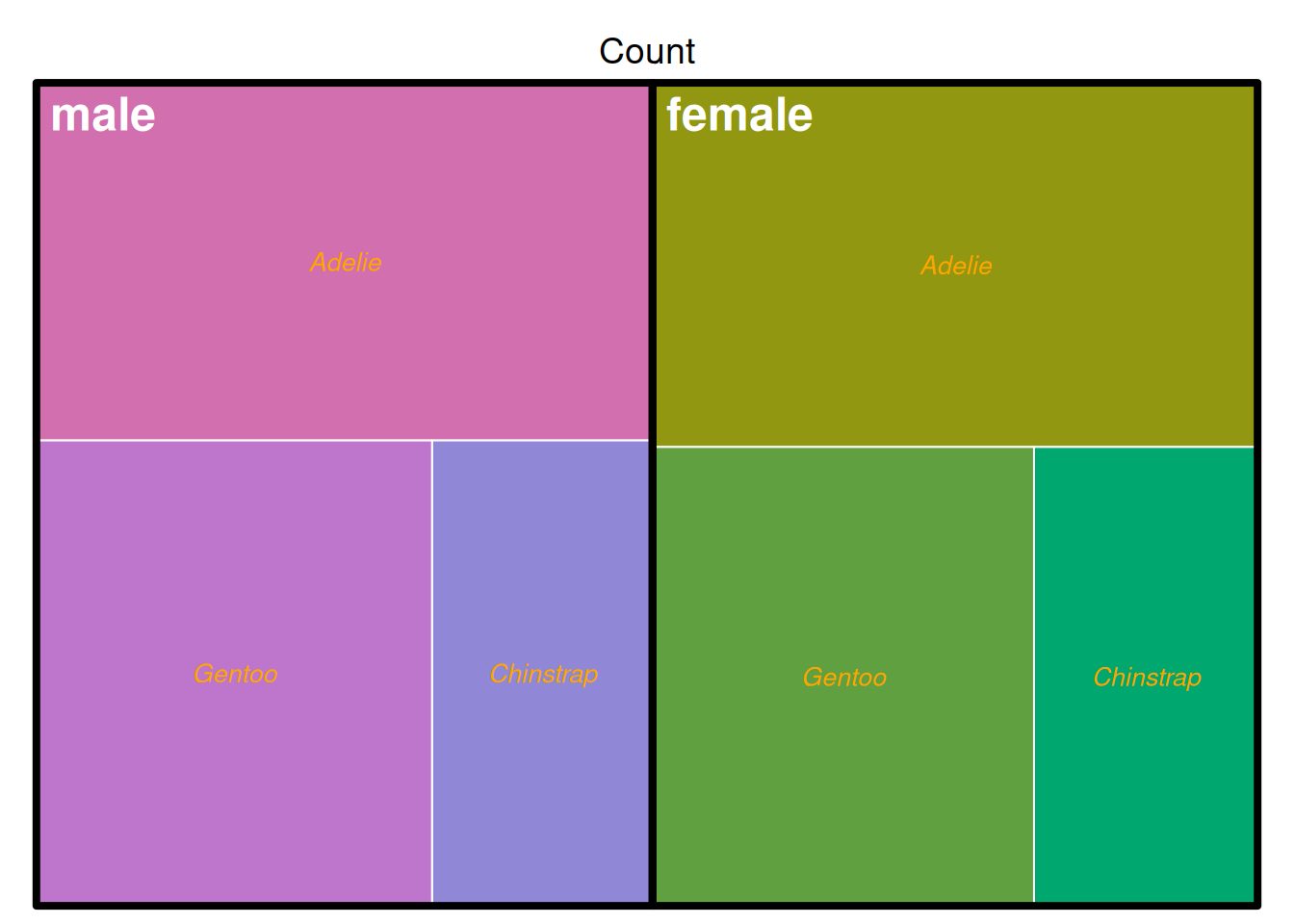
上图展示了penguins数据集中企鹅以性别与种类为依据分组的计数,上图以分类作为填色依据。
2.2 依据数据大小填色
以penguins数据集为例
treemap(data_countsub,
index = c("sex","species"),
vSize = "count",
vColor="count",
type = "value",
title = 'Count',
border.col = c("black","white"),
border.lwds = c(4,1),
fontsize.labels = c(18,10),
bg.labels=c("transparent"),
fontcolor.labels = c('white',"orange"),
align.labels = list(c("left", "top"),
c("center", "center")),
fontface.labels = c(2,3))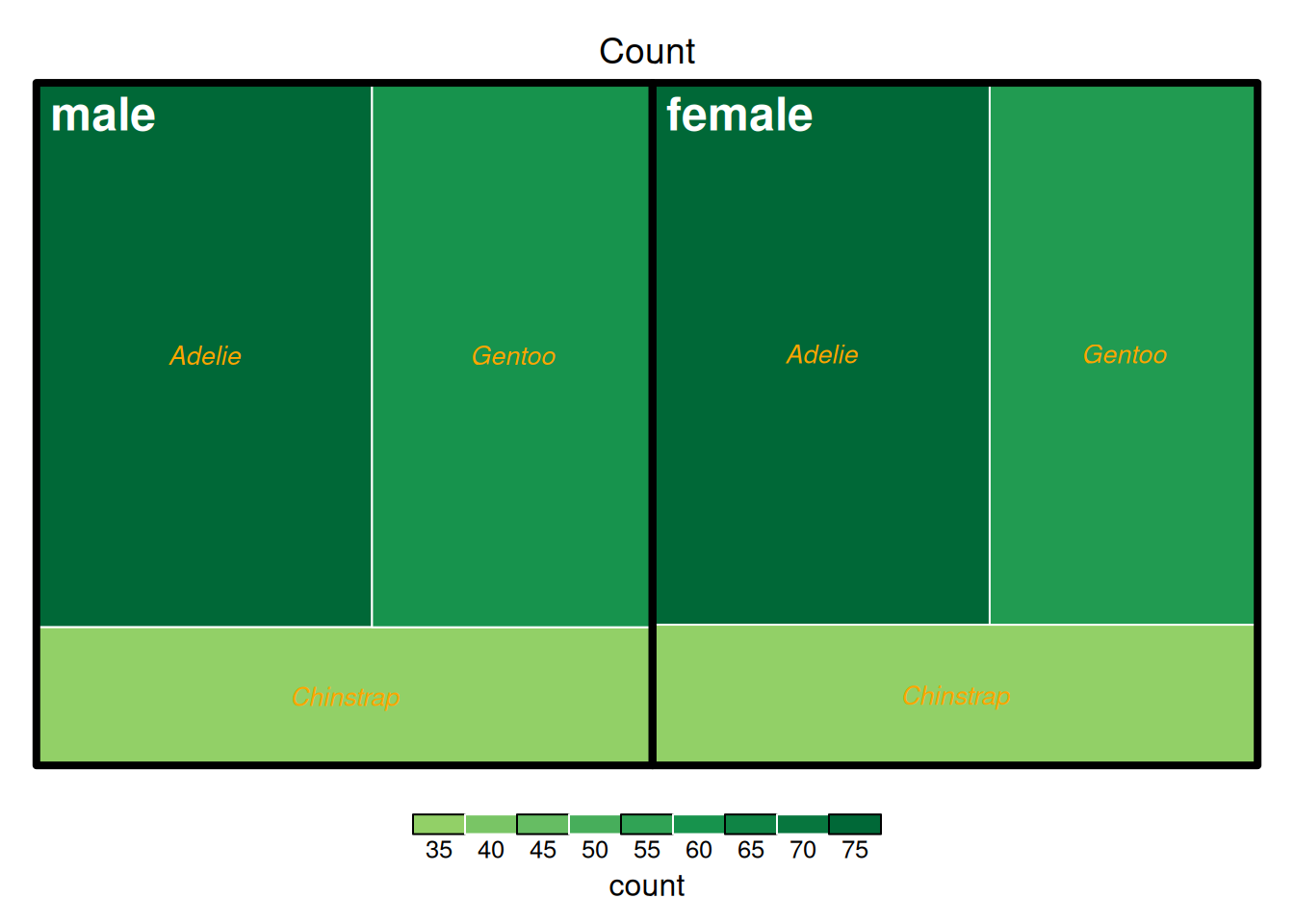
上图展示了penguins数据集中企鹅以性别与种类为依据分组的计数,上图以计数的多少作为填色依据。
以小鼠肺组织使用顺铂前后的差异基因对应的通路分析结果数据为例
treemap(data_KEGG_type,
index = c("type","subtype","PW"),
vSize = "pvalue_log",
vColor="NES",
type = "value",
title = 'KEGG通路分析结果',
border.col = c("black","white","orange"),
border.lwds = c(4,1),
fontsize.labels = c(18,15,10),
bg.labels=c("transparent"),
fontcolor.labels = c('black',"white","orange"),
align.labels = list(c("left", "top"),
c("center", "center"),
c("left","bottom")),
fontface.labels = c(2,3))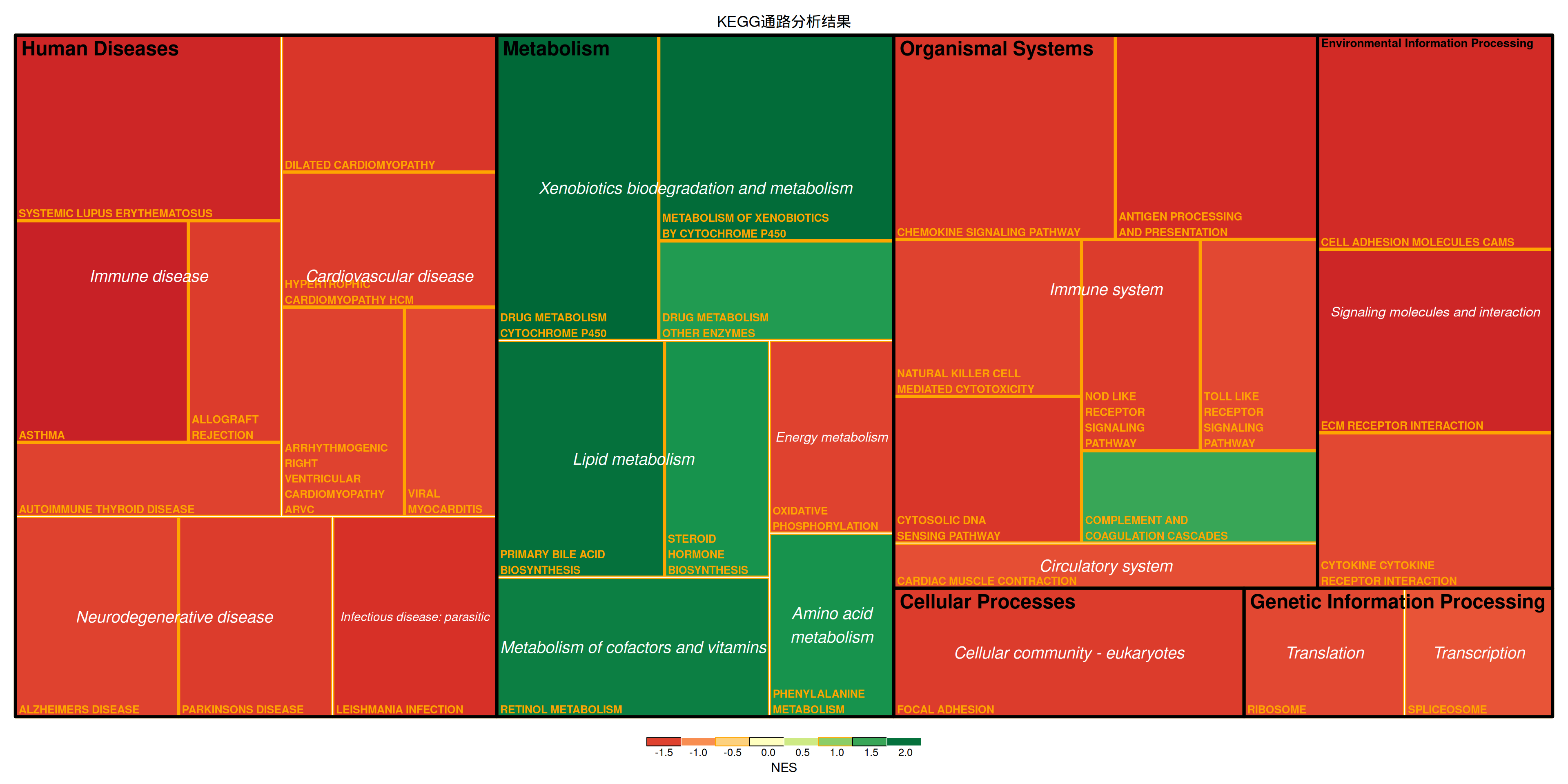
上图展示了小鼠肺组织使用顺铂前后的差异基因对应的KEGG通路分析结果。上图展示了通路的多级分类,矩形大小表示log10(P)的负值,矩形越大表示结果越可靠。矩形颜色表示NES值。
应用场景
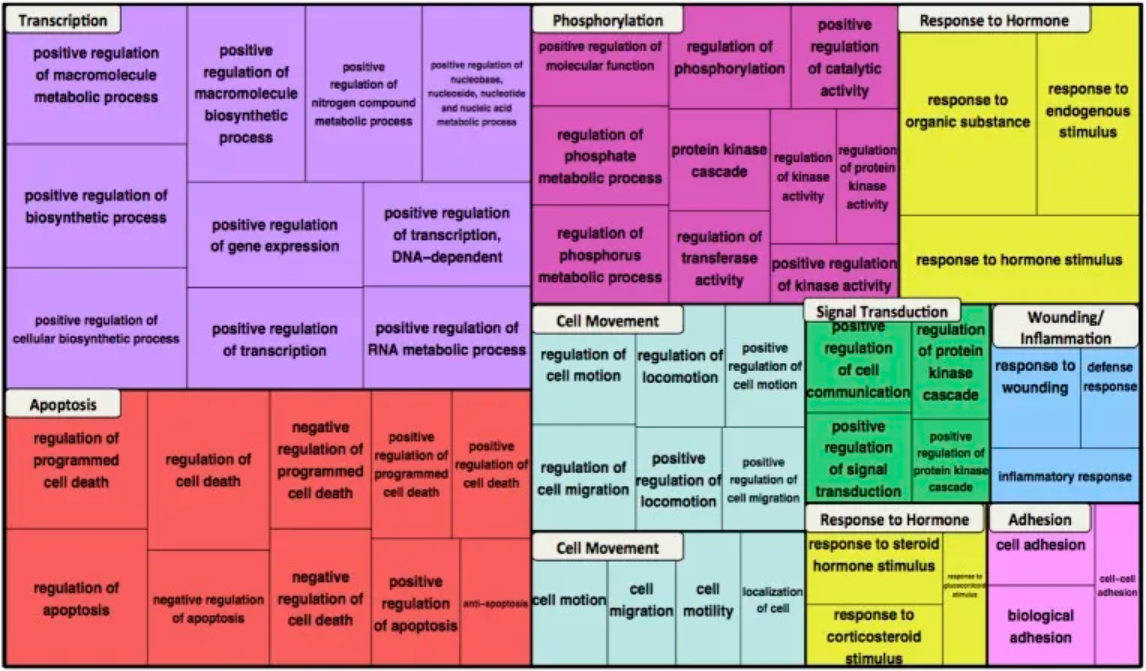
上图为β-连环蛋白网络的GO生物过程项的功能富集。前十个集群中最丰富的项显示在树图中,以不同的颜色块表示。对于每个项,方框大小反映了项浓缩的p值。[1]

上图为上调和下调基因的基因分析。使用BiNGO对上调和下调基因进行基因分析,将存在论项和相关的更正p值传递给REVIGO,REVIGO通过删除多余的GO项来执行总结。
生成与(a)上调和(b)下调基因相关的功能注释的树图。[2]
参考文献
[1] Çelen İ, Ross KE, Arighi CN, Wu CH. Bioinformatics Knowledge Map for Analysis of Beta-Catenin Function in Cancer. PLoS One. 2015 Oct 28;10(10):e0141773. doi: 10.1371/journal.pone.0141773. PMID: 26509276; PMCID: PMC4624812.
[2] Corley SM, Canales CP, Carmona-Mora P, Mendoza-Reinosa V, Beverdam A, Hardeman EC, Wilkins MR, Palmer SJ. RNA-Seq analysis of Gtf2ird1 knockout epidermal tissue provides potential insights into molecular mechanisms underpinning Williams-Beuren syndrome. BMC Genomics. 2016 Jun 13;17:450. doi: 10.1186/s12864-016-2801-4. PMID: 27295951; PMCID: PMC4907016.
[3] Tennekes M (2023). treemap: Treemap Visualization. R package version 2.4-4, https://CRAN.R-project.org/package=treemap.
[4] Wickham H, Averick M, Bryan J, Chang W, McGowan LD, François R, Grolemund G, Hayes A, Henry L, Hester J, Kuhn M, Pedersen TL, Miller E, Bache SM, Müller K, Ooms J, Robinson D, Seidel DP, Spinu V, Takahashi K, Vaughan D, Wilke C, Woo K, Yutani H (2019). “Welcome to the tidyverse.” Journal of Open Source Software, 4(43), 1686. doi:10.21105/joss.01686 https://doi.org/10.21105/joss.01686.
[5] R Core Team (2024). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
