# 安装包
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("ggpubr", quietly = TRUE)) {
install.packages("ggpubr")
}
if (!requireNamespace("patchwork", quietly = TRUE)) {
install.packages("patchwork")
}
if (!requireNamespace("dplyr", quietly = TRUE)) {
install.packages("dplyr")
}
# 加载包
library(ggplot2)
library(ggpubr)
library(patchwork)
library(dplyr)棒棒糖图
示例
(图片来源于Unsplash的Amy Shamblen)
是否对千篇一律的柱状图感到厌倦呢?如果没有蛀牙的话,不如让我们一边吃一根棒棒糖,一边将柱状图变为棒棒图吧
环境配置
系统要求: 跨平台(Linux/MacOS/Windows)
编程语言:R
依赖包:
ggplot2,ggpubr,patchwork,dplyr
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-04
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
dplyr * 1.1.4 2023-11-17 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
ggpubr * 0.6.2 2025-10-17 [1] RSPM
patchwork * 1.3.2 2025-08-25 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────数据准备
数据来源于Yang等人的文章。[1-2]
# 读取数据
data <- read.csv('https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/lollipop_1.csv', row.names = 1) #相关性分析数据读取
# 查看数据集
head(data) cell pvalue cor
1 Plasma cells 3.6e-08 0.43
2 Eosinophils 1.1e-06 0.38
3 B cells naive 5.8e-05 0.32
4 Macrophages M0 9.4e-04 0.26
5 NK cells activated 2.2e-03 0.24
6 T cells CD8 1.3e-02 0.20# 读取数据
data_2 <- read.csv('https://bizard-1301043367.cos.ap-guangzhou.myqcloud.com/lollipop_2.csv') # 基因富集分析数据读取
# 查看数据集
head(data_2) GO.category GO.Process.description..term. GO.Term.ID
1 GO:MF enzyme binding GO:0019899
2 GO:MF protein binding GO:0005515
3 GO:MF guanyl-nucleotide exchange factor activity GO:0005085
4 GO:MF Ras guanyl-nucleotide exchange factor activity GO:0005088
5 GO:MF GTPase binding GO:0051020
6 GO:MF Rho guanyl-nucleotide exchange factor activity GO:0005089
p.value Number.of.all.known.genes.enriched.to.the.GO.term
1 2.2252e-09 1,518
2 1.7950e-08 6,853
3 4.1462e-07 187
4 6.2395e-07 118
5 1.2078e-05 424
6 1.2377e-05 76
DEGs.with.GO.annotation Number.of.DEGs.enriched.to.the.particular.GO.term
1 313 71
2 313 194
3 313 20
4 313 16
5 313 28
6 313 12
Gene.names.engaged.in.particular.GO.term
1 ADCY6,ALS2,ARHGAP33,ARHGAP44,ARHGEF11,ARHGEF17,ARHGEF19,ARHGEF39,ARRDC1,AXIN2,BRD4,CAVIN3,CCNF,CLEC16A,COL1A1,CUL9,DBF4B,DENND6B,DIAPH1,DIO2,DLG4,DOCK6,DPP4,EGFR,FARP2,FURIN,GAS8,GBF1,HDAC6,HDAC7,HERC2,HTT,ISG15,KANSL1,KNDC1,KSR1,MAST2,MID1,MLPH,NCOR2,NOTCH1,OBSCN,PLEKHG2,PLEKHG3,PLK1,PPP6R2,PSD2,PTPN23,RAB11FIP3,RAPGEF1,RAPGEFL1,RBBP6,RHOF,RNF40,RPTOR,SBF1,SCAF1,SGSM1,SGSM2,SH2B1,SIK1,SKI,SLC9A1,SPATA13,SRCIN1,STRN4,TP73,TRAF3,TRIO,TSPAN5,VAV2
2 ADCY6,AKAP17A,ALOX15,ALS2,ANKRD11,ARHGAP17,ARHGAP33,ARHGAP39,ARHGAP44,ARHGEF11,ARHGEF17,ARHGEF19,ARHGEF39,ARNT2,ARRDC1,ATXN1L,AXIN2,BCL9L,BRD3,BRD4,BTBD19,BTNL9,C12H17ORF100,C1QA,C1QTNF6,CACNA1G,CAMSAP1,CASKIN1,CAVIN3,CCNF,CD59,CD79A,CDH24,CENPT,CEP131,CEP164,CEP170B,CHD2,CHD7,CHERP,CLEC16A,COL1A1,COL5A1,CREBBP,CRSP3,CRTC1,CSRP1,CUL9,DBF4B,DENND4B,DENND6B,DGKD,DIAPH1,DIO2,DLG4,DLL1,DNAH1,DOCK6,DPP4,DYNC1H1,E2F1,E2F7,EFS,EGFR,EHBP1L1,EHMT1,ELMSAN1,ENSSSCG00000035856,EP300,FARP2,FBXL18,FURIN,FXYD1,GAS7,GAS8,GBF1,GIGYF1,GIPC3,HDAC6,HDAC7,HERC2,HTT,IGSF9,INPPL1,ISG15,KANSL1,KCNQ4,KCP,KCTD7,KIF12,KIF18B,KIF1A,KIF1C,KIF21B,KIF26A,KIF7,KMT2B,KNDC1,KSR1,LARP1,LDB3,LRP5,LZTS1,MAST2,MED12,MEGF8,MID1,MLPH,MMRN1,MNT,MRAP2,MYH14,MYH3,MYH7B,MYH9,NAV2,NCOR2,NECTIN1,NOTCH1,NSD2,NUMA1,OBSCN,PAX2,PER3,PIK3R2,PLCG1,PLEKHA6,PLEKHG2,PLEKHG3,PLK1,PLXNA1,PLXNA3,PLXNB3,PPP6R2,PRDM15,PSD2,PTPN23,PTPRF,RAB11FIP3,RAPGEF1,RAPGEFL1,RBBP6,RERE,RGS2,RHOF,RNF123,RNF40,RPTOR,RSPO3,S100A9,SALL1,SARM1,SART3,SBF1,SCAF1,SEMA4C,SEMA4F,SETD5,SGSM1,SGSM2,SH2B1,SH3PXD2B,SIK1,SKI,SLC9A1,SPATA13,SPTAN1,SRCIN1,STRN4,SYNE2,SYT3,TCAP,TENM2,TLE2,TMEFF2,TONSL,TP73,TRAF3,TRANK1,TRAPPC12,TRIM66,TRIO,TRRAP,TSPAN5,TSPO,TYK2,UBR4,UNC5B,USP20,VAV2,VEGFA,WDFY3,WDR62,ZMYND8
3 ALS2,ARHGEF11,ARHGEF17,ARHGEF19,ARHGEF39,DENND6B,DOCK6,FARP2,GBF1,KNDC1,OBSCN,PLEKHG2,PLEKHG3,PSD2,RAPGEF1,RAPGEFL1,SBF1,SPATA13,TRIO,VAV2
4 ALS2,ARHGEF11,ARHGEF17,ARHGEF19,ARHGEF39,DENND6B,FARP2,KNDC1,OBSCN,PLEKHG2,PLEKHG3,RAPGEF1,SBF1,SPATA13,TRIO,VAV2
5 ALS2,ARHGAP44,ARHGEF11,ARHGEF17,ARHGEF19,ARHGEF39,CLEC16A,DENND6B,DIAPH1,DOCK6,FARP2,GAS8,GBF1,KNDC1,MLPH,OBSCN,PLEKHG2,PLEKHG3,PSD2,RAB11FIP3,RAPGEF1,RAPGEFL1,SBF1,SGSM1,SGSM2,SPATA13,TRIO,VAV2
6 ALS2,ARHGEF11,ARHGEF17,ARHGEF19,ARHGEF39,FARP2,OBSCN,PLEKHG2,PLEKHG3,SPATA13,TRIO,VAV2可视化
1. 基础绘图
基础棒棒图显示了COL17A1基因与免疫浸润相关性的结果。[1]
# 基础棒棒图
# 对相关系数和p值转换为分类变量
data$pvalue_group <- cut(data$pvalue,
breaks = c(0, 0.2, 0.4, 0.6,0.8, 1),
labels = c("< 0.2","< 0.4","< 0.6","< 0.8","<1"),
right=FALSE)# right=FALSE表示表示区间为左闭右开
data$cor_group_size <- cut(abs(data$cor),# 绝对值
breaks = c(0, 0.1, 0.2, 0.3, 0.4, 0.5),
labels = c("0.1","0.2","0.3","0.4","0.5"),
right=FALSE)
# 排序
data = data[order(data$cor),]
data$cell = factor(data$cell, levels = data$cell)
p = ggplot(data,
aes(x = cor, y = cell, color = pvalue_group)) +
scale_color_manual(name="pvalue",
values = c("#146432",
"#4DB748",
#"#FAA519", #由于没有这个区间的数据,故注释掉
"#FABECD" #,
#"#FAD700"#由于没有这个区间的数据,故注释掉
))+ #棒棒糖中糖果的颜色选择
geom_segment(aes(x = 0, y = cell, xend = cor, yend = cell),
color = 'black', #棒棒糖中棒子的绘制
linewidth = 0.5) +
geom_point(aes(size = cor_group_size))+ #棒棒糖中糖果的绘制
labs(title = "COL17A1",#图片标题
size = "abs(cor)") + #图例的名称
guides(color = "none")+ #隐藏多余的图例
theme_bw()+ #清空格式
theme(plot.title=element_text(size=8, #标题大小
hjust=0.5 ), #标题位置
legend.position = "bottom", #图例的位置
text = element_text(family = "serif"),#设置字体为Times New Roman
panel.grid = element_line(linetype = "dotted",color='grey'))
p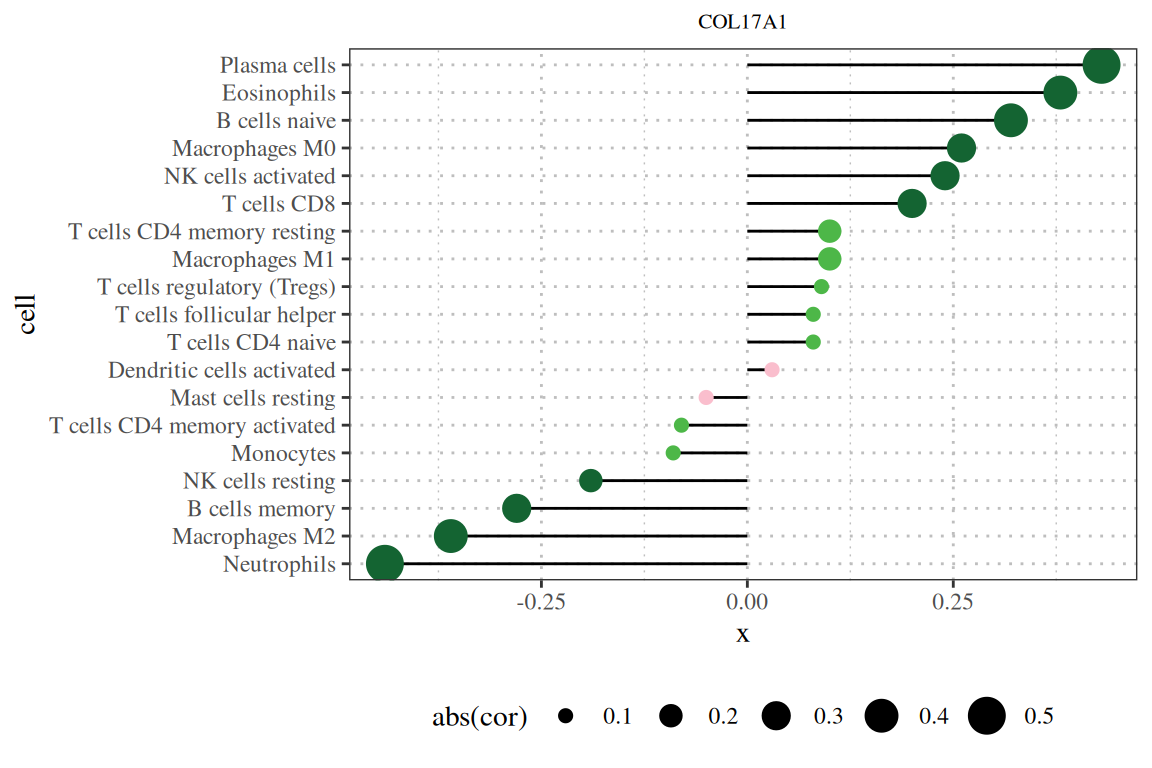
注:图题为基因名,纵轴为系报名,横轴为该基因的表达量,cor为基因表达量与细胞丰度的相关性,其中横轴为相关性,大小为相关性的绝对值,颜色为P值。
为了使棒棒糖图展示的信息更丰富,我们可以在图右侧添加信息,我们在上一步已经依据p值进行了排序,在这一步可以增加p值的文本,使得图片可读性更强。
data$pvalue_col <- cut(data$pvalue,
breaks = c(0, 0.05,1),
labels = c("< 0.05","> 0.05"),
right=FALSE) #在data中添加颜色分类信息
data$pvalue_text <- ifelse(data$pvalue>0.05,sprintf("%.3f", data$pvalue),'<0.001')#在data中添加需要绘制的文本
p1 = ggplot()+
geom_text(data,mapping = aes(x = 0, y = cell, color = pvalue_col,
label = pvalue_text))+
scale_color_manual(name="",values = c("red", "black"))+
theme_void()+
theme(text = element_text(family = "serif"))+
guides(color=F) #去掉图例
p|p1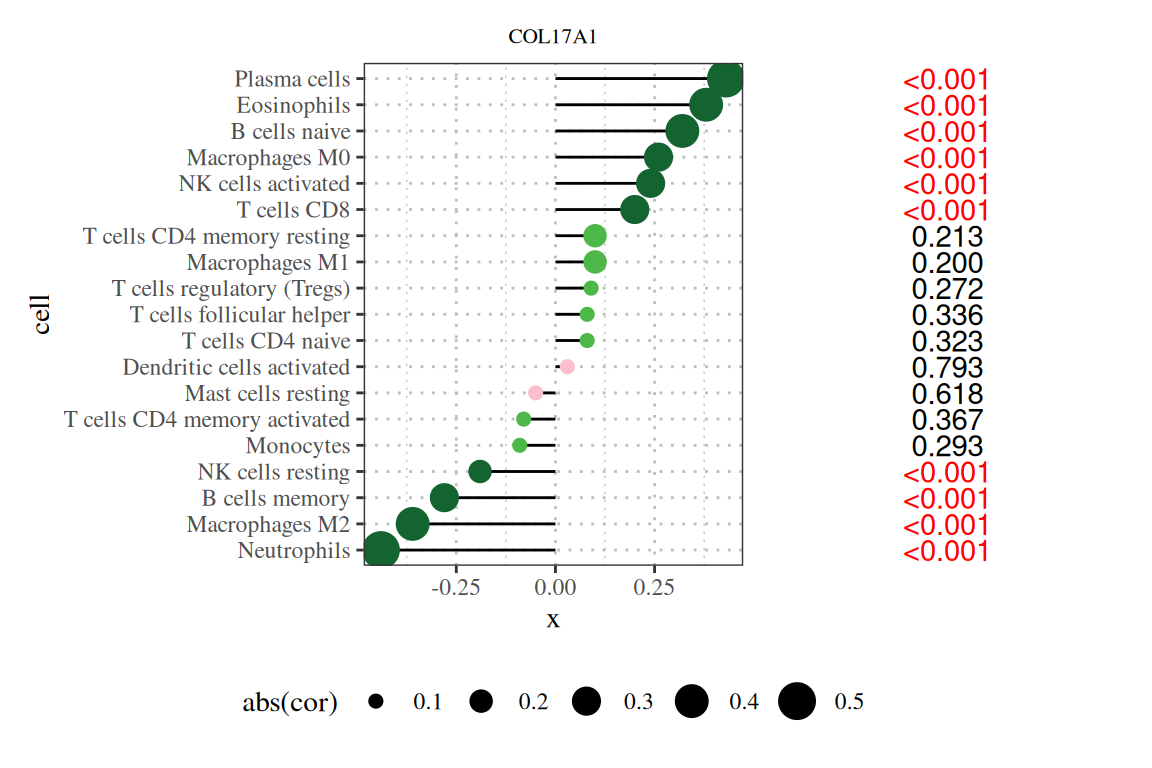
注:图题为基因名,纵轴为系报名,横轴为该基因的表达量,cor为基因表达量与细胞丰度的相关性,其中横轴为相关性,大小为相关性的绝对值,颜色为P值,右侧数字为P值。
2. 添加图例
stack_data = data.frame( x = c("legend","legend","legend","legend","legend"),
class = c("0-0.2", "0.2-0.4", "0.4-0.6", "0.6-0.8", "0.8-1"),
color_range = c(0.2,0.2,0.2,0.2,0.2))
p2 <- ggplot(stack_data, aes(x = x, y = color_range, fill = class)) +
geom_bar(stat = 'identity', position = "stack", width = 0.3) +
scale_fill_manual(name = "P-value",values = c("#146432", "#4DB748", "#FAA519", "#FABECD", "#FAD700")) + # 设置颜色
scale_y_continuous(breaks = seq(0, 1, by = 0.2),
labels = c("0","0.2","0.4","0.5","0.8","1.0" ),
sec.axis = sec_axis(~., breaks = seq(0, 1, by = 0.2),
labels = c("0", "0.2", "0.4", "0.6", "0.8", "1.0"))) + # 设置y轴刻度并将y轴移动到右侧
labs(title = "pvalue")+#图片标题
theme_minimal() +
theme(axis.text.x = element_blank(), # 去掉x轴标签
axis.title.x = element_blank(), # 去掉x轴标题
axis.text.y = element_blank(), # 去掉y轴左侧标题
axis.title.y = element_blank(), # 去掉y轴左侧标题
panel.grid = element_blank(), # 去掉背景网格线
plot.margin = unit(c(0, 0, 0, 0), "cm"), # 去掉图形的四周空白
axis.text.y.right = element_text(size = 12), # 设置y轴右侧标签的字体大小
legend.position = "none", # 去掉图例
plot.title=element_text(size=10, #标题大小
hjust=0.5 ), #标题位置
text = element_text(family = "serif")) #设置字体为Times New Roman
p|p1|p2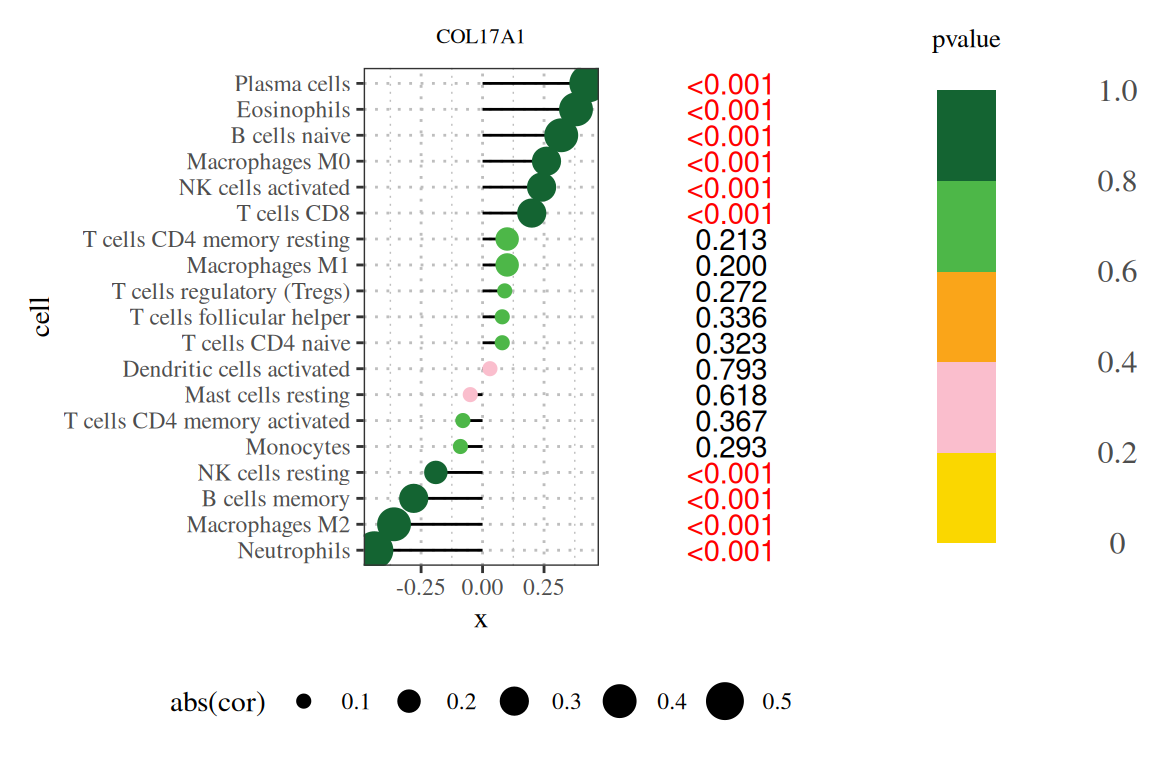
注:图题为基因名,纵轴为系报名,横轴为该基因的表达量,cor为基因表达量与细胞丰度的相关性,其中横轴为相关性,大小为相关性的绝对值,颜色为P值,右侧数字为P值。
3. 美化拼图
layout <- c(
area(t = 1, l = 1, b = 1, r = 2),
area(t = 1, l = 3, b = 1, r = 3),
area(t = 1, l = 4, b = 1, r = 4)
)
p + p1 + p2 +
plot_layout(design = layout)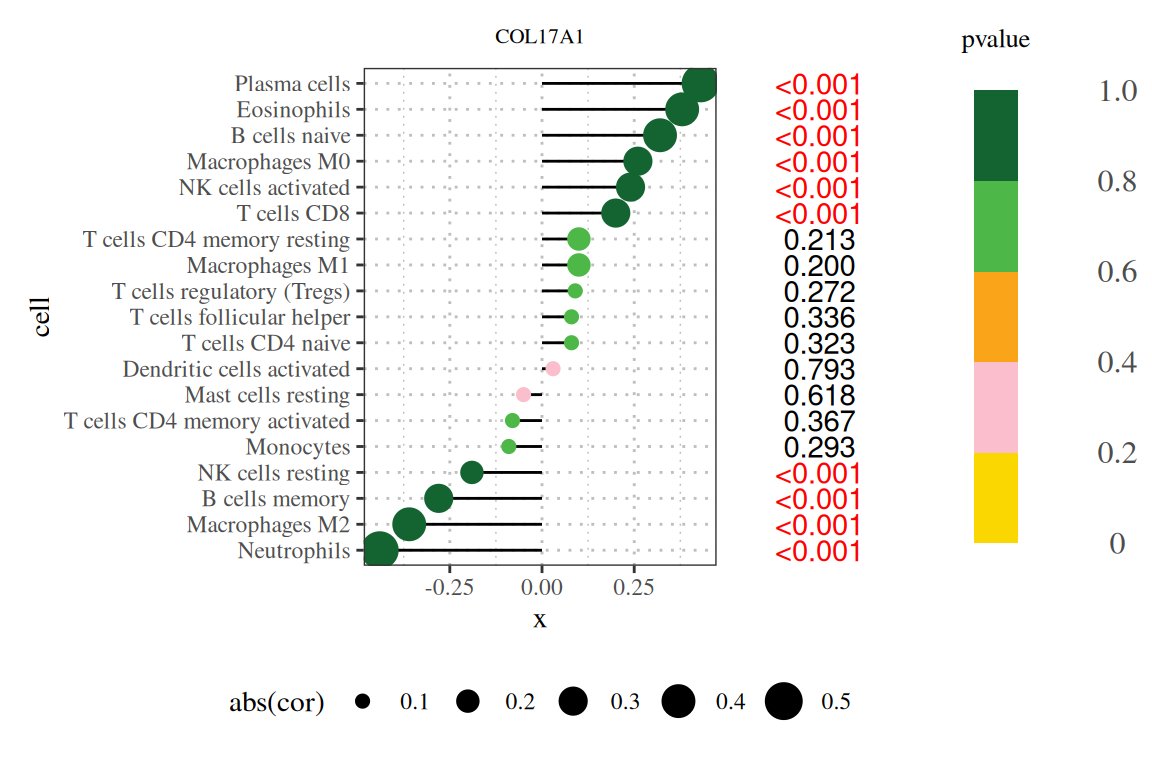
如果有需要的话可以进一步使用Power point或者Adobe illustration进行美化。
4. 富集分析绘图
# 富集分析棒棒糖图
data_2 <- data_2[,c(1,2,4,7)] #只留下绘图需要的信息
colnames(data_2) <- c("GO_aspect","GO_term","P","count") #重命名列名
data_2 <- data_2[data_2$GO_aspect!='KEGG',] #去除掉KEGG富集分析的数据
data_2$Padj <- p.adjust(data_2$P, method = "BH")# 使用Benjamin-Hochberg方法计算Padj
data_2$log10Padj = -log10(data_2$Padj)#计算-log10Padj
data_2$GO_aspect[data_2$GO_aspect=="GO:BP"] ="BP"
data_2$GO_aspect[data_2$GO_aspect=="GO:CC"] ="CC"
data_2$GO_aspect[data_2$GO_aspect=="GO:MF"] ="MF"
# 按GO_aspect分组,然后每组按log10Padj排序,并取前15条数据
data_2 <- data_2 %>% group_by(GO_aspect) %>%
arrange(GO_aspect, desc(log10Padj)) %>% # 按log10Padj降序排序
slice_head(n = 15) # 取前15条
#绘图
ggdotchart(data_2, x = "GO_term", y = "log10Padj",
color = "GO_aspect", # 按组显示颜色
palette = c("#090886", "#F90708", "#25821F"), # 自定义调色板
sorting = "descending", # 按降序对值排序
add = "segments", # 添加从y=0到点的线段
group = "GO_aspect", # 按组排序
dot.size = 8, # 点大小
label = round(data_2$count), # 将mpg值添加为点标签
font.label = list(color = "white", size = 9,
vjust = 0.5), # 调整标签参数
ggtheme = theme_pubr() # ggplot2主题
)+labs(x=NULL,y=expression(-log[10](Padj)),
title=NULL)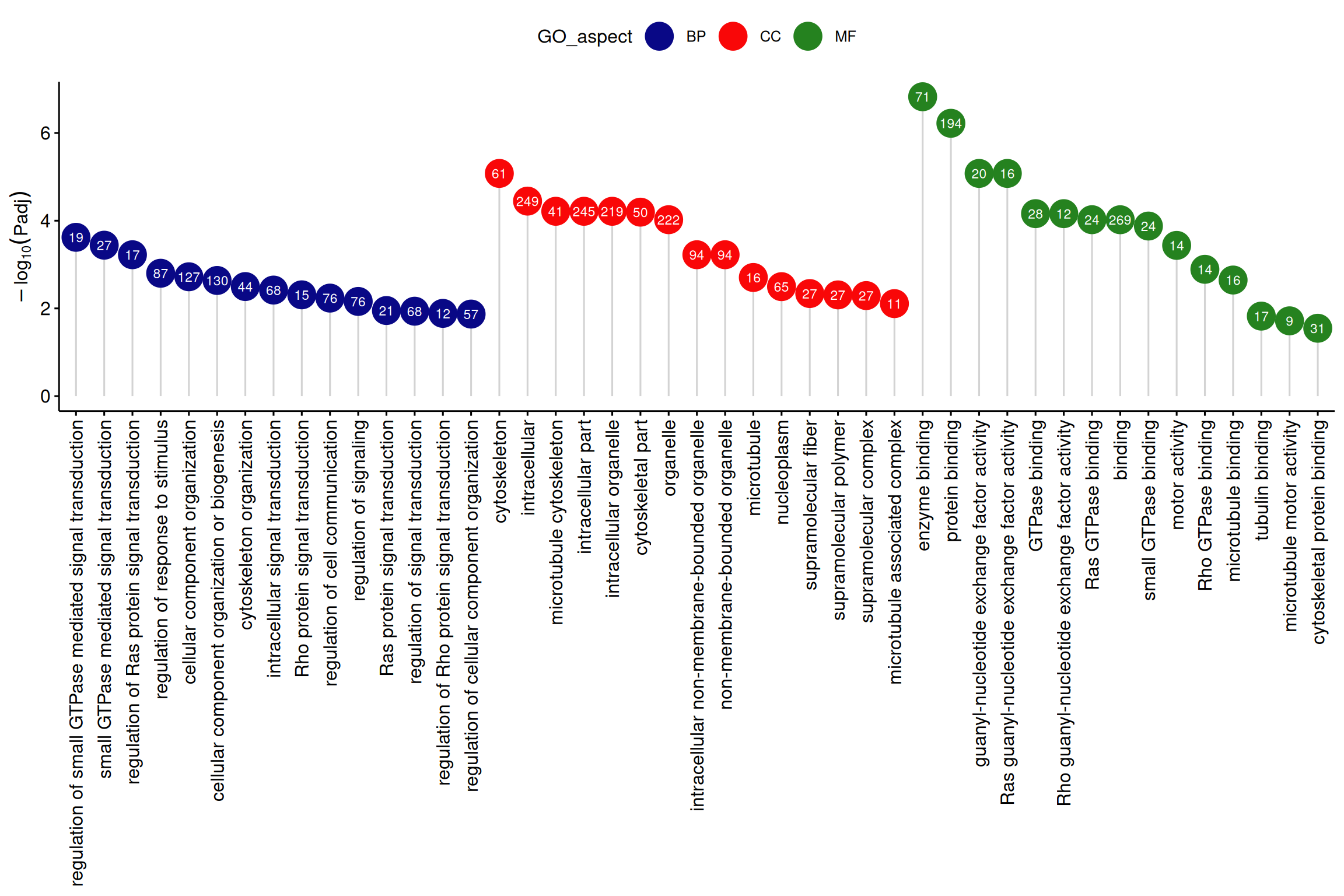
注:纵轴为-log10Padj,横轴为GO条目,颜色为GO类别,圆圈中数字为富集到各GO条目的基因个数。
应用场景
1. 相关性分析
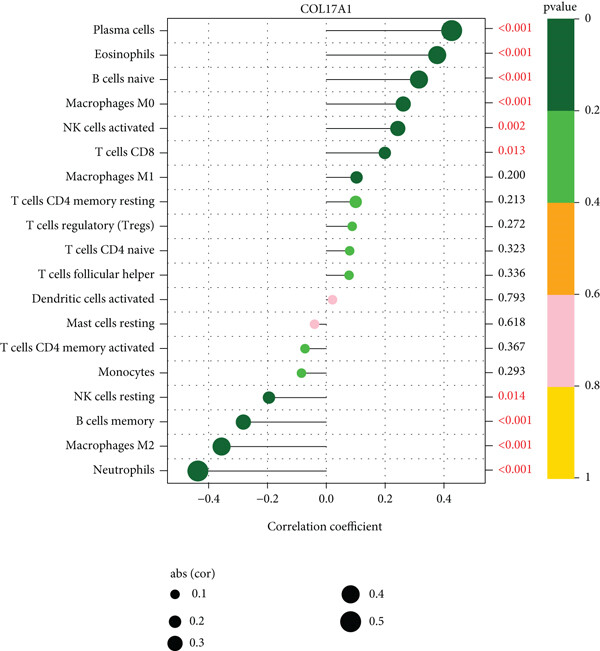
棒棒糖图(原文图9A)的应用展示了COL17A1基因与免疫浸润相关性的结果。[1]
2. 基因通路富集分析
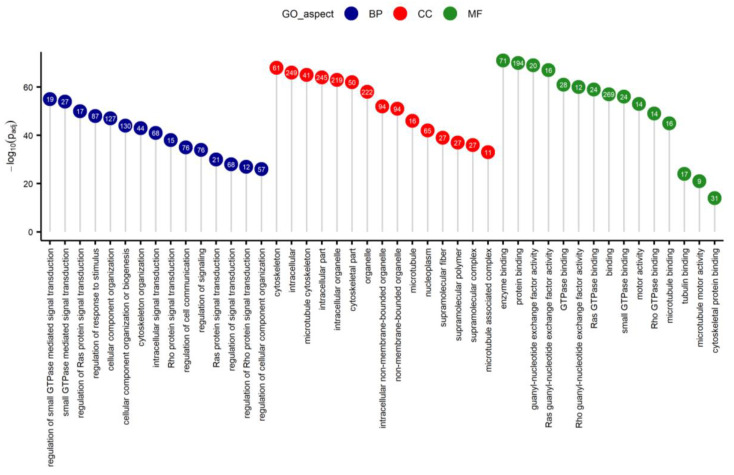
棒棒糖图(原文图3)的应用展示了差异表达基因的GO富集分析结果。[2]
参考文献
[1] Yang, M. Y., Ji, M. H., Shen, T., & Lei, L. (2022). Integrated Analysis Identifies Four Genes as Novel Diagnostic Biomarkers Which Correlate with Immune Infiltration in Preeclampsia. Journal of immunology research, 2022, 2373694. https://doi.org/10.1155/2022/2373694
[2] Paukszto, L., Mikolajczyk, A., Jastrzebski, J. P., Majewska, M., Dobrzyn, K., Kiezun, M., Smolinska, N., & Kaminski, T. (2020). Transcriptome, Spliceosome and Editome Expression Patterns of the Porcine Endometrium in Response to a Single Subclinical Dose of Salmonella Enteritidis Lipopolysaccharide. International journal of molecular sciences, 21(12), 4217. https://doi.org/10.3390/ijms21124217
