# Install packages
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("grafify", quietly = TRUE)) {
install.packages("grafify")
}
# Load packages
library(data.table)
library(jsonlite)
library(grafify)QQ Plot
Note
Hiplot website
This page is the tutorial for source code version of the Hiplot QQ Plot plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
Verify whether a set of data comes from a certain distribution or whether two sets of data come from the same (family) distribution.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
data.table;jsonlite;grafify
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-03
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.2.1 2026-01-27 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
grafify * 5.1.0 2025-08-25 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────Data Preparation
# Load data
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/qqplot/data.json")$exampleData[[1]]$textarea[[1]])
data <- as.data.frame(data)
# Convert data structure
data[, "Genotype"] <- factor(data[, "Genotype"], levels = unique(data[, "Genotype"]))
# View data
head(data) Genotype Cytokine Experiment facet
1 WT 1.312322 Exp_ 1 A
2 WT 2.171466 Exp_ 2 A
3 WT 1.497610 Exp_ 3 A
4 WT 1.088861 Exp_ 4 A
5 WT 1.270528 Exp_ 5 A
6 WT 3.012708 Exp_ 6 AVisualization
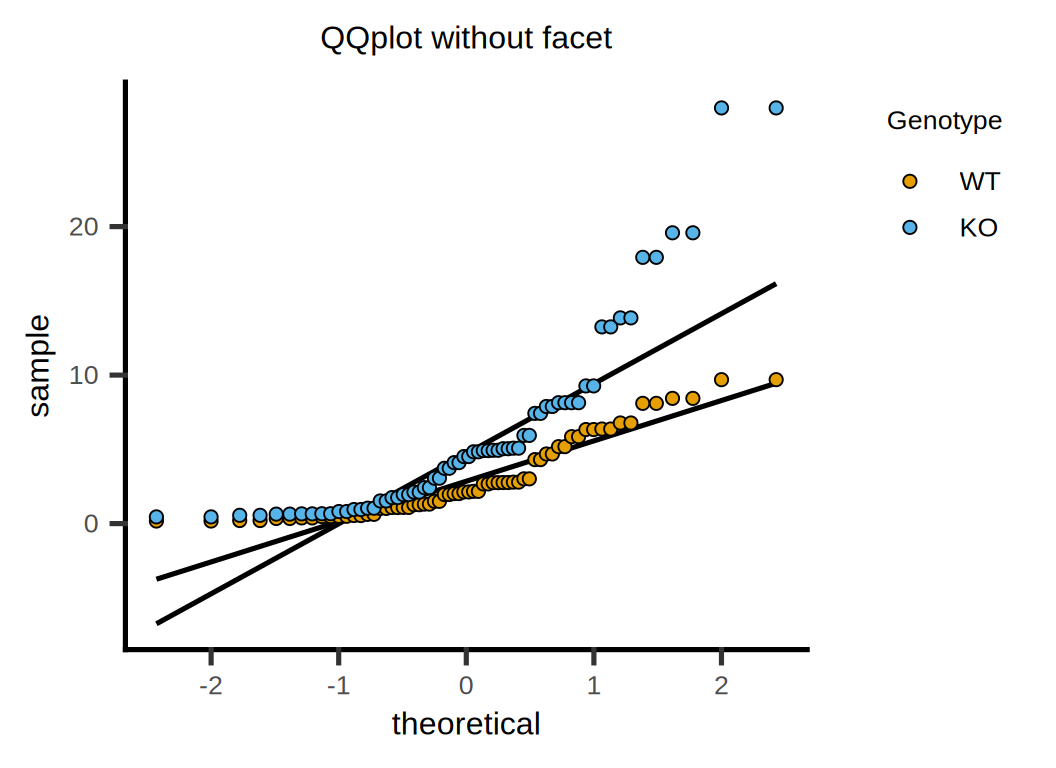
# QQ Plot
p <- plot_qqline(data = data, ycol = Cytokine, group = Genotype,
symsize = 2, symthick = 0.5, s_alpha = 1) +
ggtitle("QQplot without facet") +
xlab("theoretical") + ylab("sample") +
guides(fill = guide_legend(title = "Genotype")) +
scale_color_manual(values = c("#E69F00","#4DB1DC")) +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p