# Install packages
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("rms", quietly = TRUE)) {
install.packages("rms")
}
if (!requireNamespace("survival", quietly = TRUE)) {
install.packages("survival")
}
if (!requireNamespace("ggplot2", quietly = TRUE)) {
install.packages("ggplot2")
}
if (!requireNamespace("stringr", quietly = TRUE)) {
install.packages("stringr")
}
# Load packages
library(data.table)
library(jsonlite)
library(rms)
library(survival)
library(ggplot2)
library(stringr)RCS-LRM
Note
Hiplot website
This page is the tutorial for source code version of the Hiplot RCS-LRM plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
Nonlinear regression analysis.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
data.table;jsonlite;rms;survival;ggplot2;stringr
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-01-27
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
data.table * 1.18.0 2025-12-24 [1] RSPM
ggplot2 * 4.0.1 2025-11-14 [1] RSPM
Hmisc * 5.2-5 2026-01-09 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
rms * 8.1-0 2025-10-14 [1] RSPM
stringr * 1.6.0 2025-11-04 [1] RSPM
survival * 3.8-3 2024-12-17 [3] CRAN (R 4.5.2)
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────Data Preparation
# Load data
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/rcs-lrm/data.json")$exampleData$textarea[[1]])
data <- as.data.frame(data)
# Convert data structure
data <- na.omit(data)
ex <- set::not(colnames(data), c("main", "group"))
ex <- str_c(ex, collapse = "+")
dd <<- datadist(data)
options(datadist = "dd")
for (i in 3:5) {
fit <- lrm(as.formula(paste0("group~rcs(main,nk=i,inclx = T)+", ex, collapse = "+")), data = data, x = TRUE)
tmp <- AIC(fit)
if (i == 3) {
AIC <- tmp
nk <<- 3
}
if (tmp < AIC) {
AIC <- tmp
nk <<- i
}
}
fit <- lrm(as.formula(paste0("group~rcs(main,nk=nk,inclx = T)+", ex, collapse = "+")), data = data, x = TRUE)
dd$limits$main[2] <- median(data$main)
fit <- update(fit)
orr <- Predict(fit, main, fun = exp, ref.zero = TRUE)
# View data
head(data) main X2 X3 group
1 100 0.90 0 1
2 90 0.65 1 1
3 400 1.36 0 1
4 200 0.83 0 1
5 300 1.38 0 1
6 200 0.69 0 1Visualization
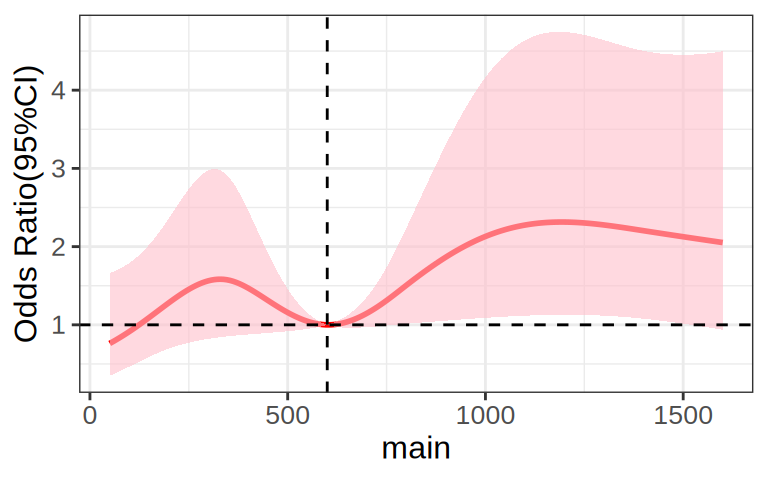
# RCS-LRM
p <- ggplot() +
geom_line(data = orr, aes(main, yhat), linetype = "solid", size = 1, alpha = 1,
colour = "#FF0000") +
geom_ribbon(data = orr, aes(main, ymin = lower, ymax = upper), alpha = 0.6,
fill = "#FFC0CB") +
geom_hline(yintercept = 1, linetype = 2, size = 0.5) +
geom_vline(xintercept = dd$limits$main[2], linetype = 2, size = 0.5) +
labs(x = "main", y = "Odds Ratio(95%CI)") +
theme_bw() +
theme(text = element_text(family = "Arial"),
plot.title = element_text(size = 12,hjust = 0.5),
axis.title = element_text(size = 12),
axis.text = element_text(size = 10),
axis.text.x = element_text(angle = 0, hjust = 0.5,vjust = 1),
legend.position = "right",
legend.direction = "vertical",
legend.title = element_text(size = 10),
legend.text = element_text(size = 10))
p