# Install packages
if (!requireNamespace("data.table", quietly = TRUE)) {
install.packages("data.table")
}
if (!requireNamespace("jsonlite", quietly = TRUE)) {
install.packages("jsonlite")
}
if (!requireNamespace("sigminer", quietly = TRUE)) {
remotes::install_github("ShixiangWang/sigminer")
}
# Load packages
library(data.table)
library(jsonlite)
library(sigminer)Group-comparison Heatmap
Note
Hiplot website
This page is the tutorial for source code version of the Hiplot Group-comparison Heatmap plugin. You can also use the Hiplot website to achieve no code ploting. For more information please see the following link:
Group-comparison Heatmap provides a way to compare multiple variables across multiple (>2) groups and visualize the result with heatmap.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
data.table;jsonlite;sigminer
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-03
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
Biobase * 2.70.0 2025-10-29 [1] Bioconduc~
BiocGenerics * 0.56.0 2025-10-29 [1] Bioconduc~
data.table * 1.18.2.1 2026-01-27 [1] RSPM
generics * 0.1.4 2025-05-09 [1] RSPM
jsonlite * 2.0.0 2025-03-27 [1] RSPM
sigminer * 2.3.1 2024-05-11 [1] RSPM
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────Data Preparation
# Load data
data <- data.table::fread(jsonlite::read_json("https://hiplot.cn/ui/basic/group-comparison/data.json")$exampleData$textarea[[1]])
data <- as.data.frame(data)
# View data
head(data) g1 g2 e1 e2
1 A AA N -1.8060313
2 A AA N -0.5820759
3 A AA N -1.1088896
4 A AA N -1.0149620
5 A AA P -0.1623095
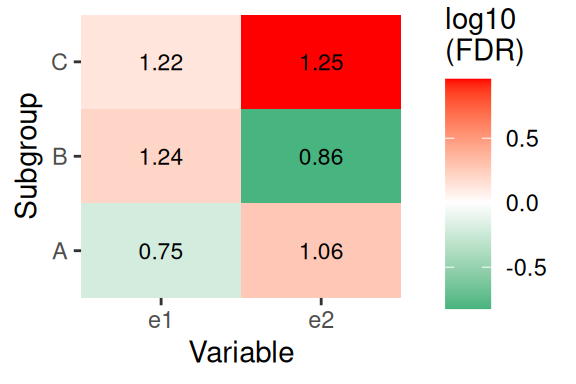
6 A AA N 0.5630558Visualization
# Define plot functions
unlist_and_covert <- function(x, recursive = FALSE) {
if (!is.null(x)) {
x <- unlist(x, recursive = recursive)
if (!is.null(x)) {
y <- sapply(x, function(x) {
if (identical(x, "NA")) NA else x
})
names(y) <- names(x)
x <- y
}
}
x
}
plotentry <- function(data,
grp_vars = NULL, enrich_vars = NULL, cross = TRUE,
co_method = c("t.test", "wilcox.test"), ref_group = NA,
scales = "free", add_text_annotation = TRUE,
fill_by_p_value = TRUE, use_fdr = TRUE, cut_p_value = FALSE,
cluster_row = FALSE) {
ref_group <- unlist_and_covert(ref_group)
if (is.null(ref_group)) ref_group <- NA
rv <- group_enrichment(data, grp_vars, enrich_vars, cross, co_method, ref_group)
if (length(unique(rv$grp_var)) == 1) {
p <- show_group_enrichment(rv,
return_list = TRUE,
scales = scales, add_text_annotation = add_text_annotation,
fill_by_p_value = fill_by_p_value, use_fdr = use_fdr, cut_p_value = cut_p_value,
cluster_row = cluster_row
)
p <- p[[1]]
} else {
p <- show_group_enrichment(rv,
scales = scales, add_text_annotation = add_text_annotation,
fill_by_p_value = fill_by_p_value, use_fdr = use_fdr, cut_p_value = cut_p_value,
cluster_row = cluster_row
)
}
return(p)
}
# plot
p <- plotentry(
data = data,
grp_vars = "g1",
enrich_vars = c("e1", "e2"),
cross = T,
add_text_annotation = T,
fill_by_p_value = T,
use_fdr = T,
cut_p_value = F,
cluster_row = F,
co_method = "t.test",
scales = "free"
)
p