# Installing packages
if (!requireNamespace("survival", quietly = TRUE)) {
install.packages("survival")
}
if (!requireNamespace("gtsummary", quietly = TRUE)) {
install.packages("gtsummary")
}
if (!requireNamespace("dplyr", quietly = TRUE)) {
install.packages("dplyr")
}
if (!requireNamespace("datawizard", quietly = TRUE)) {
install.packages("datawizard")
}
if (!requireNamespace("broom.helpers", quietly = TRUE)) {
remotes::install_github("larmarange/broom.helpers")
}
# Load packages
library(survival)
library(gtsummary)
library(dplyr)
library(datawizard)
library(broom.helpers)Regression Analysis Table
The regression analysis table is used to display the results of the regression model. It provides statistical information about the variables in the model and helps explain the relationship between the variables.
Example
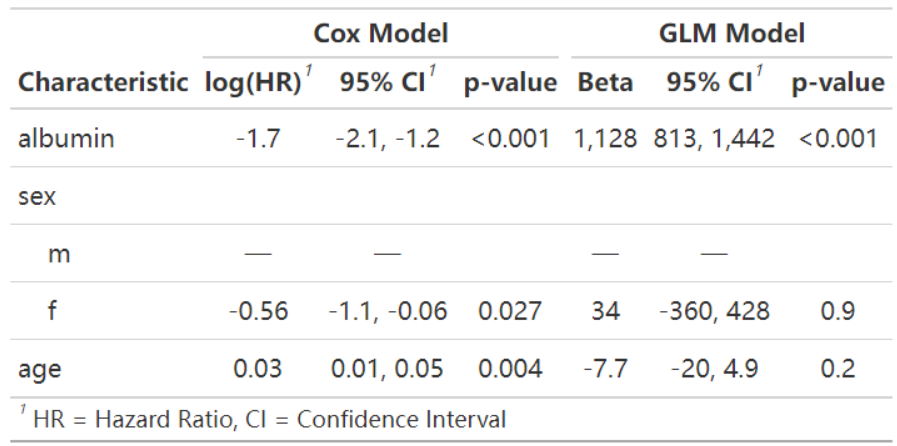
The figure below shows a regression analysis based on the pbc dataset built into the survival package. The figure uses the Cox proportional hazards model and generalized linear regression model to explore the effects of serum protein, sex, and age on survival.
Setup
System Requirements: Cross-platform (Linux/MacOS/Windows)
Programming language: R
Dependent packages:
survival,gtsummary,dplyr,datawizard
sessioninfo::session_info("attached")─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.5.2 (2025-10-31)
os Ubuntu 24.04.3 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate C.UTF-8
ctype C.UTF-8
tz UTC
date 2026-02-01
pandoc 3.1.3 @ /usr/bin/ (via rmarkdown)
quarto 1.8.27 @ /usr/local/bin/quarto
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
broom.helpers * 1.22.0.9000 2025-11-02 [1] Github (larmarange/broom.helpers@02687ca)
datawizard * 1.3.0 2025-10-11 [1] RSPM
dplyr * 1.1.4 2023-11-17 [1] RSPM
gtsummary * 2.5.0 2025-12-05 [1] RSPM
survival * 3.8-3 2024-12-17 [3] CRAN (R 4.5.2)
[1] /home/runner/work/_temp/Library
[2] /opt/R/4.5.2/lib/R/site-library
[3] /opt/R/4.5.2/lib/R/library
* ── Packages attached to the search path.
──────────────────────────────────────────────────────────────────────────────Data Preparation
The pbc dataset, built into the survival R package, contains information on 418 patients with primary biliary cirrhosis (PBC) who had received ursodeoxycholic acid (UDCA) treatment at or before the start of the study. The pbc dataset records clinical information such as survival time, survival status, treatment method, age, sex, albumin level, and hepatomegaly.
df <- pbc %>%
filter(status != 1) %>%
mutate(status = ifelse(status == 2, 1, 0)) %>%
select(2:13) %>%
na.omit() %>%
# Divide `albumin` into 3 groups
mutate(albumin3cat = categorize(albumin, split = "quantile", n_groups = 3))
head(df[,1:6]) time status trt age sex ascites
1 400 1 1 58.76523 f 1
2 4500 0 1 56.44627 f 0
3 1012 1 1 70.07255 m 0
4 1925 1 1 54.74059 f 0
5 2503 1 2 66.25873 f 0
6 1832 0 2 55.53457 f 0Visualization
1. Basic regression analysis table
# Basic regression analysis table
t1 <- coxph(Surv(time, status) ~ albumin + sex + age,
data = df
) %>%
tbl_regression()
t1| Characteristic | log(HR) | 95% CI | p-value |
|---|---|---|---|
| albumin | -1.7 | -2.1, -1.2 | <0.001 |
| sex | |||
| m | — | — | |
| f | -0.56 | -1.1, -0.06 | 0.027 |
| age | 0.03 | 0.01, 0.05 | 0.004 |
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | |||
This table is a basic regression analysis table. The Cox proportional hazard model is called by coxph() and the results of the regression analysis are tabulated using tbl_regression().
2. tbl_regression() parameter settings
# tbl_regression() parameter settings
t1 <- coxph(Surv(time, status) ~ albumin + sex + age,
data = df
) %>%
# parameter settings
tbl_regression(
conf.level = 0.90,
exponentiate = TRUE,
include = c("sex", "albumin"),
show_single_row = sex,
label = list(sex ~ "sex as categorical")
)
t1| Characteristic | HR | 90% CI | p-value |
|---|---|---|---|
| sex as categorical | 0.57 | 0.37, 0.87 | 0.027 |
| albumin | 0.19 | 0.13, 0.28 | <0.001 |
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | |||
There are many parameter settings in the table tbl_regression(), which can change the confidence interval, row names and other information of the table.
Important parameter: tbl_regression
conf.level: Determines the confidence level for the regression analysis. The default value of 0.95 indicates a 95% confidence interval.exponentiate: Whether to exponentiate the HR values. The default column is log(HR) values.include: Which independent variables (rows) are included in the statistical table.show_single_row: Applies to binary variables and does not display the control group.label: Changes the name (row name) of the independent variable.
3. Add global-p
# Add global-p
df$albumin3cat=as.factor(df$albumin3cat)
t1 <- coxph(Surv(time, status) ~ albumin3cat + sex + age,
data = df) %>%
tbl_regression() %>%
add_global_p()
t1| Characteristic | log(HR) | 95% CI | p-value |
|---|---|---|---|
| albumin3cat | <0.001 | ||
| 1 | — | — | |
| 2 | -0.72 | -1.2, -0.27 | |
| 3 | -1.3 | -1.7, -0.77 | |
| sex | 0.073 | ||
| m | — | — | |
| f | -0.48 | -0.98, 0.02 | |
| age | 0.03 | 0.01, 0.05 | <0.001 |
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | |||
This table adds global P values via add_global_p().
4. Add q-value
# Add q-value
t1 <- coxph(Surv(time, status) ~ albumin + sex + age,
data = df
) %>%
tbl_regression() %>%
# Add a q-value column
add_q()
t1| Characteristic | log(HR) | 95% CI | p-value | q-value1 |
|---|---|---|---|---|
| albumin | -1.7 | -2.1, -1.2 | <0.001 | <0.001 |
| sex | ||||
| m | — | — | ||
| f | -0.56 | -1.1, -0.06 | 0.027 | 0.027 |
| age | 0.03 | 0.01, 0.05 | 0.004 | 0.006 |
| 1 False discovery rate correction for multiple testing | ||||
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | ||||
The table adds a q-value column via add_q().
5. Merge columns
# Merge columns
t1 <- coxph(Surv(time, status) ~ albumin3cat + sex + age,
data = df) %>%
tbl_regression() %>%
# Modify the header name
modify_header(p.value = "**P**",estimate="**log(HR) (CI)**") %>%
# Combine the score column and the CI column and apply to rows where the score is not NA
modify_column_merge(
pattern = "{estimate} ({conf.low}, {conf.high})",
rows = !is.na(estimate)
)
t1| Characteristic | log(HR) (CI) | P |
|---|---|---|
| albumin3cat | ||
| 1 | — | |
| 2 | -0.72 (-1.2, -0.27) | 0.002 |
| 3 | -1.3 (-1.7, -0.77) | <0.001 |
| sex | ||
| m | — | |
| f | -0.48 (-0.98, 0.02) | 0.062 |
| age | 0.03 (0.01, 0.05) | <0.001 |
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | ||
This table merges the score column and the CI column, which is a common form. The table header is appropriately modified after the merger.
Important Functions modify_header / modify_column_merge:
modify_header:modify_header() can be used to modify the header name. label represents the variable column, p.value represents the P-value column, ci represents the confidence interval column, estimate represents the score column, conf.low represents the lower confidence interval, and conf.high represents the upper confidence interval.
modify_column_merge: The pattern parameter specifies the pattern of columns to be merged, and the rows parameter specifies the rows to apply the merge to.
6. Model integration
Model horizontal integration合
# Model horizontal integration
t1 <- coxph(Surv(time, status) ~ albumin + sex + age,
data = df
) %>%
tbl_regression()
t2 <- glm(time ~ albumin + sex + age,
data = df
) %>%
tbl_regression()
# t1, t2 model integration
t3 <- tbl_merge(
tbls = list(t1, t2),
tab_spanner = c("**Cox Model**", "**GLM Model**")
)
t3| Characteristic |
Cox Model
|
GLM Model
|
||||
|---|---|---|---|---|---|---|
| log(HR) | 95% CI | p-value | Beta | 95% CI | p-value | |
| albumin | -1.7 | -2.1, -1.2 | <0.001 | 1,128 | 813, 1,442 | <0.001 |
| sex | ||||||
| m | — | — | — | — | ||
| f | -0.56 | -1.1, -0.06 | 0.027 | 34 | -360, 428 | 0.9 |
| age | 0.03 | 0.01, 0.05 | 0.004 | -7.7 | -20, 4.9 | 0.2 |
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | ||||||
This table uses tbl_merge to horizontally integrate the results of the Cox proportional hazards model and the generalized linear regression model for patients with pdb.
Model vertical integration
# Model vertical integration
t1 <- coxph(Surv(time, status) ~ albumin + sex + age,
data = df[df$hepato==0,]
) %>%
tbl_regression()
t2 <- coxph(Surv(time, status) ~ albumin + sex + age,
data = df[df$hepato==1,]
) %>%
tbl_regression()
tbl_stack(list(t1, t2), group_header = c("Patients without hepato", "Patients with hepato"))| Characteristic | log(HR) | 95% CI | p-value |
|---|---|---|---|
| Patients without hepato | |||
| albumin | -1.4 | -2.3, -0.47 | 0.003 |
| sex | |||
| m | — | — | |
| f | -1.1 | -1.9, -0.36 | 0.004 |
| age | 0.06 | 0.03, 0.09 | <0.001 |
| Patients with hepato | |||
| albumin | -1.4 | -2.0, -0.87 | <0.001 |
| sex | |||
| m | — | — | |
| f | -0.29 | -0.97, 0.38 | 0.4 |
| age | 0.01 | -0.01, 0.04 | 0.3 |
| Abbreviations: CI = Confidence Interval, HR = Hazard Ratio | |||
This table uses tbl_stack to longitudinally integrate the results of the Cox proportional hazards model for the two groups of pdb patients with and without hepato.
Application
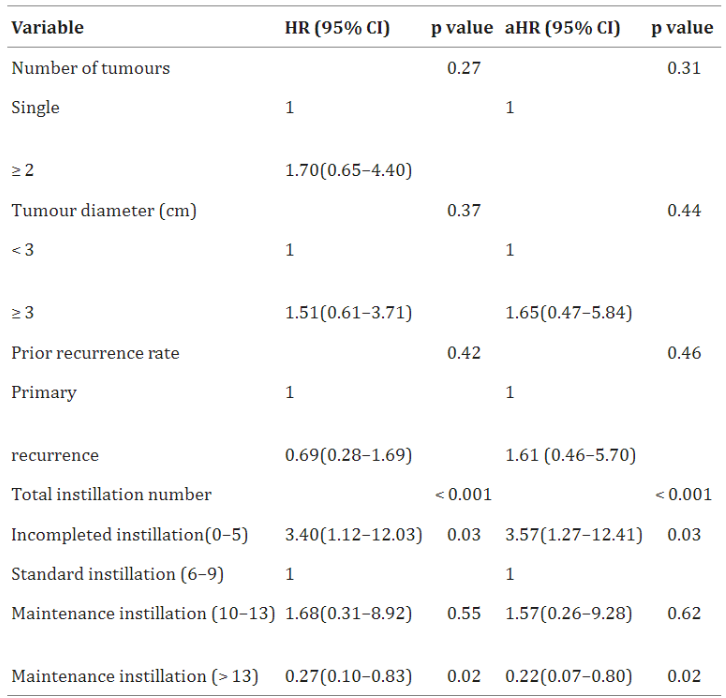
Univariate and multivariate Cox regression analyses were performed to assess 3-year tumor recurrence in patients with intermediate-risk NMIBC. [1]
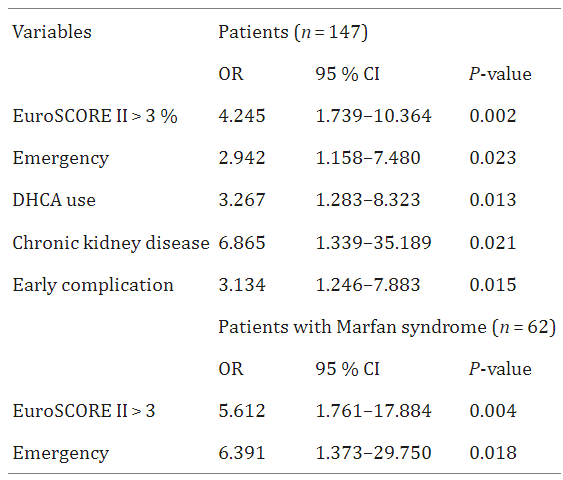
This regression analysis table shows the independent predictors of late total mortality in multivariable Cox regression analysis. [2]
Reference
[1] CHEN J X, HUANG W T, ZHANG Q Y, et al. The optimal intravesical maintenance chemotherapy scheme for the intermediate-risk group non-muscle-invasive bladder cancer[J]. BMC Cancer, 2023,23(1): 1018.
[2] BENKE K, ÁGG B, SZABÓ L, et al. Bentall procedure: quarter century of clinical experiences of a single surgeon[J]. J Cardiothorac Surg, 2016,11: 19.
